Abstract
Communication between cells ensures coordinated behavior. In prokaryotes, this signaling is typically referred to as quorum sensing, whereas in eukaryotic cells, communication occurs through hormones. In recent years, reports have shown that small noncoding RNAs, called microRNAs (miRNAs), can be transmitted from one species to another, inducing signal interference in distant species, even in a cross-kingdom manner. This new mode of cross-species communication might mediate symbiotic and pathogenic relationships between various organisms (e.g., microorganisms and their hosts). Here, we discuss several recent studies concerning miRNA-mediated cross-species gene regulation.
Introduction
Organisms do not exist in isolation; instead, most organisms communicate through interconnected ecosystems. The research field of cross-species communication has received significant attention in recent years. This concept is defined as the communication between different species of animals, plants, fungi and bacteria through an array of hormones or hormone-like chemicals. This field evolved from the initial concept in the early 1980s that different cell types can communicate with each other through secreted hormones.Citation1 Subsequently, this field expanded with the realizations that bacterial signals modulate mammalian and plant cell signal transduction and that host hormones also cross-signal with quorum-sensing molecules to modulate bacterial gene expression.Citation2-Citation4
Recently, scientists have discovered an entirely new level of gene regulation mediated through small molecules called microRNAs (miRNAs). miRNAs are a class of single-stranded noncoding RNAs of approximately 22 nucleotides in length that play an important role in post-transcriptional gene regulation.Citation5,Citation6 It is now accepted that miRNAs play crucial regulatory roles in numerous biological processes, including cell proliferation, differentiation, development, apoptosis and metabolism.Citation7 Strikingly, recent findings have suggested that miRNAs not only execute functions within the original cells but can also be transmitted from one species to another, facilitating cross-talk, communication or signal interference between different species, even in a cross-kingdom manner. The identification of this novel mechanism has greatly enhanced our understanding of molecular signaling between species. Here, we discuss several recent studies concerning miRNA-mediated cross-species gene regulation.
Plant miRNAs regulate gene expression in mammals
Our recent studies utilizing oxidized deep sequencing and quantitative RT-PCR revealed that plant miRNAs bearing 2’-O-methylated 3′ ends could stably exist in the sera and tissues of humans and herbivorous animals at concentrations similar to some endogenous mammalian miRNAs.Citation8 Furthermore, using a murine model, we demonstrated that plant miRNAs accumulate in the serum and tissues as a result of food intake and that exogenous mature plant miRNAs in food pass through the mouse gastrointestinal tract.Citation8 Moreover, we showed that, in mammals, low-density lipoprotein receptor adaptor protein 1 (LDLRAP1) is a target of MIR168a, which is one of the plant miRNAs present at relatively high levels in human serum, and MIR168a directly binds to the coding sequence of LDLRAP1 in human liver cells and influences the uptake of LDL from the blood.Citation8 This finding provides evidence that food-derived exogenous plant miRNAs enter into the circulation and organs of mammals, where they serve functions similar to endogenous miRNAs, simultaneously regulating multiple target genes and biological processes.
However, the uptake of plant-derived miRNAs into the body through food sources and eventual absorption into the target organs remains challenging. Once inside the mammalian gastrointestinal tract, exogenous miRNAs encounter RNases, phagocytosis and a low-pH environment, unfavorable conditions that might require the miRNAs to adopt stable structures for protection against degradation prior to reaching the recipient cells. However, the mechanism underlying the absorption of exogenous plant miRNAs into intestinal epithelial cells remains unclear. It is also unknown how plant miRNAs are eventually delivered to the cells of target organs, such as the liver in mammals, after uptake. Thus, it is essential to characterize the mechanisms underlying the absorption and processing of exogenous plant miRNAs in mammalian cells.
Human miRNAs affect malaria parasite biology and survival
Recently, LaMonte et al. reported that human miRNAs could be translocated into the malaria parasite Plasmodium falciparum, where they are incorporated covalently into P. falciparum mRNAs to repress mRNA translation, leading to a modest but significant reduction in the growth of P. falciparum.Citation9 Specifically, these authors detected the presence of 100 human miRNAs within the parasites using multiplex real-time PCR assays. Interestingly, some human miRNAs, which are abundant in HbAS and HbSS erythrocytes (e.g., miR-451), were significantly enriched in the parasite.Citation9 Using northern blot analyses, these authors verified that the intra-parasitic miRNAs were fully intact mature human miRNAs and ruled out the possibility that the parasitic miRNAs are derived from host erythrocyte contamination.Citation9 To validate the miRNA uptake profile across the intraerythrocytic developmental cycle (IDC), LaMonte et al. examined three miRNAs and observed that let-7i was maximally expressed after 16 h of parasite growth, whereas the expression of miR-451 peaked after 32 h.Citation9 These differences strongly suggest that the uptake of miRNA might be an active, dynamic process. Moreover, miR-451 was localized within the parasitophorous vacuolar membrane (PVM).Citation9 Subsequently, using Illumina deep sequencing, 5′ RACE and ribonuclease protection assays, these authors confirmed that the human miRNAs transferred into the parasite (e.g., miR-451) formed chimeric fusions with P. falciparum mRNA and, via impaired ribosomal loading, resulted in translational inhibition, which eventually affected the parasite biology and survival.Citation9 Thus, sickle cell erythrocytes exhibit cell-intrinsic resistance to malaria in part through an atypical miRNA activity, which may represent a unique host defense strategy against complex eukaryotic pathogens.
This study was the first to show that human miRNAs regulate protozoan gene expression and provides an explanation for the parasite resistance typically observed in the red blood cell disorder β-thalassemia. However, this study also raises some questions. First, it is currently unknown how miRNAs are incorporated into malaria parasites. Plasmodium parasites are contained within a parasitophorous vacuolar membrane; the miRNA would have to cross through both the red blood cell and parasite plasma membranes. Thus, it will be of great interest to elucidate the mechanism of miRNA translocation, and the direct visualization of miR-451 activity in the parasitophorous vacuolar membrane should be reconciled. Second, Plasmodium parasites do not contain Dicer or Argonaute homologs that comprise the RNA-induced silencing complex (RISC). RNA interference, including the traditional function of miRNAs, is not observed in these organisms. Thus, it is not surprising that translocated miRNAs are covalently linked to certain Plasmodium mRNAs and form chimeric RNAs, independent of the conventional role of miRNAs in post-transcriptional gene regulation. Covalent linkage requires a trans-splicing event; however, among unicellular eukaryotes, trans-splicing has only been observed in kinetoplastid organisms, whereby a leader sequence is spliced onto the 5′ end of all RNA transcripts during processing and maturation.Citation10 In P. falciparum, miRNA tagging occurs for a minority of transcripts, including regulatory subunit of cAMP-dependent protein kinase (PKA-R), phosphoethanolamine N-methyltransferase (PEAMT) and 28S and 18S rRNAs. The mechanism that determines the specific enrichment of certain miRNAs or the incorporation of these miRNAs into specific parasite mRNAs is unknown, and whether Argonaute proteins are involved in these processes has not been determined.
The regulation of viral and host gene expression occurs through miRNAs
Another example of miRNA-mediated cross-species communication comes from virus-host interactions. Numerous studies have demonstrated that viruses hijack the host cell miRNA machinery to generate their own miRNAs.Citation11-Citation13 Although the functions of most viral miRNAs remain unknown, current evidence indicates that viruses use miRNAs to manipulate both viral and host gene expression.Citation14-Citation21 In general, viral miRNAs target cellular genes involved in cell proliferation, survival, stress responses or antiviral defense pathways, as prolonging cell survival and evading immune recognition are at least two mechanisms by which viral miRNAs facilitate the virus life cycle.Citation16-Citation21 Thus, viruses take advantage of the host cell miRNA machinery and establish a cellular environment conducive to viral replication. More interestingly, a handful of viral miRNAs exhibit homology to human oncogenic miRNAs. For example, Kaposi’s sarcoma-associated herpesvirus (KSHV)-encoded miR-K12-11 is an ortholog of miR-155.Citation22,Citation23 One shared cellular target of miR-K12-11 and miR-155 is BACH-1 (Btb and CNC homolog 1), a transcriptional repressor involved in the oxidative stress response and cell cycle regulation.Citation22,Citation23 Given the known etiological role of miR-155 in B-cell transformation, it has been suggested that miR-K12-11 might contribute to the induction of KSHV-positive B-cell tumors in infected patients.Citation22,Citation23 Hence, viruses might utilize the molecular mimicry of miRNA genes to regulate host cellular environments in the same manner as the homologous cytokines, growth factors and immunomodulatory genes commonly found in virus genomes.
There are other mechanisms by which host miRNAs influence the life cycles of viruses. It has been shown that miR-24 and miR-93 target viral large protein (L protein) and phosphoprotein (P protein) genes in mice infected with vesicular stomatitis virus (VSV).Citation24 Similarly, a cluster of cellular miRNAs, including miR-28, miR-125b, miR-150, miR-223 and miR-382, target the 3′ ends of HIV-1 mRNAs in cultivated resting CD4+ T cells to maintain HIV-1 latency.Citation25 Moreever, it has been reported that miR-32 effectively restricts the accumulation of retrovirus primate foamy virus type 1 (PFV-1) in human cells in a direct, sequence-specific manner.Citation26 Therefore, through the fortuitous recognition of foreign nucleic acids, cellular miRNAs exert direct antiviral effects in addition to their conventional post-transcriptionally regulatory functions. Perhaps the role of miR-122 in controlling hepatitis C virus (HCV) infection is the most interesting example of the control of viruses through host miRNAs. miR-122 stimulates HCV replication through unique interactions at two binding sites in the 5′ UTR of the HCV genome.Citation27 In this context, miR-122 stabilizes viral RNA and reduces its decay rather than inducing viral RNA degradation, thereby expanding the knowledge of how miRNAs modulate gene expression in multiple ways.Citation27
Taken together, these data show that while viral miRNAs profoundly impact host gene expression to augment viral replication, host miRNAs directly interact with viruses. These findings suggest that miRNAs play an important role in the intricate virus-host interaction network, providing new insights into the diagnosis and control of viral diseases. However, compared with miRNAs transmitted from plants to mammals or from humans to plasmodia, there are major differences for miRNAs of viral origin. In the strictest sense, viruses are typically not considered to be individual organisms or living entities. Thus, virus-encoded miRNAs rely on the host machinery for production, whereas plant or human miRNAs are synthesized and subsequently transmitted to other organisms. Therefore, the miRNA-mediated cross-species communication between viruses and hosts is somewhat different from that observed in other systems.
Conclusion
Once considered unstable molecules, miRNAs have recently been shown to transfer horizontally between species, suggesting a novel role for these molecules in interspecies communication. Thus, the miRNAs endogenous to one species might influence the biology of other distantly related species ().
Figure 1. Cross-species regulation of gene expression through miRNAs. (A) Plant miRNAs (e.g., MIR168a) regulate gene expression in mammals.Citation8 (B) Human miRNAs (e.g., miR-451) affect malaria parasite biology and survival.Citation9 (C) Viral miRNAs (e.g., KSHV-miR-K12-11) regulate host gene expression,Citation16-Citation23 and host miRNAs (e.g., miR-122) regulate viral gene expression.Citation24-Citation27
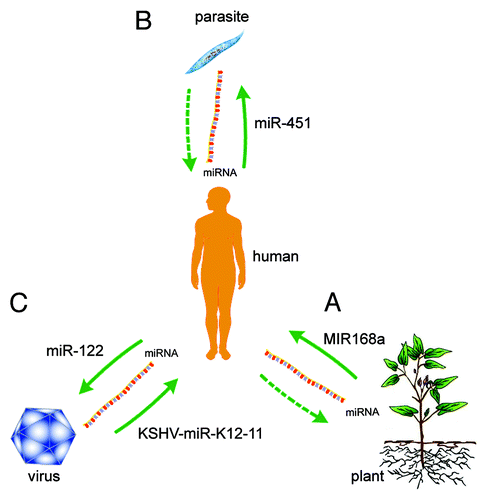
It is not surprising that viruses and hosts regulate eachothers’ gene expression patterns through miRNAs. However, for each others’ species, this regulation is more complicated. Previous studies have shown that the malaria parasite does not encode miRNAs,Citation28 yet these pathogens might encode other small non-coding RNAs. We propose that Plasmodium regulates gene expression in host cells through small non-coding RNAs. Furthermore, we have shown that animals absorb plant miRNAs from plant foods, thereby affecting their own gene expression. However, it is unknown whether plants absorb animal miRNAs from animal waste or the animal ingredients in organic fertilizers to affect plant gene expression. Thus, interspecies miRNA interactions will be a new and exciting field of study, and much remains to be learned about the mechanisms and roles of miRNAs in both normal physiological and pathological contexts before we can fully understand exogenous miRNA-mediated cross-species communication.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (Nos. 81101330, 31271378, 81250044 and J1103512) and the Natural Science Foundation of Jiangsu Province (Nos. BK2011013 and BK2012014).
References
- Roth J, LeRoith D, Shiloach J, Rosenzweig JL, Lesniak MA, Havrankova J. The evolutionary origins of hormones, neurotransmitters, and other extracellular chemical messengers: implications for mammalian biology. N Engl J Med 1982; 306:523 - 7; http://dx.doi.org/10.1056/NEJM198203043060907; PMID: 6120460
- Pacheco AR, Sperandio V. Inter-kingdom signaling: chemical language between bacteria and host. Curr Opin Microbiol 2009; 12:192 - 8; http://dx.doi.org/10.1016/j.mib.2009.01.006; PMID: 19318290
- Sperandio V. Striking a balance: inter-kingdom cell-to-cell signaling, friendship or war?. Trends Immunol 2004; 25:505 - 7; http://dx.doi.org/10.1016/j.it.2004.08.001; PMID: 15364050
- Hughes DT, Sperandio V. Inter-kingdom signalling: communication between bacteria and their hosts. Nat Rev Microbiol 2008; 6:111 - 20; http://dx.doi.org/10.1038/nrmicro1836; PMID: 18197168
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281 - 97; http://dx.doi.org/10.1016/S0092-8674(04)00045-5; PMID: 14744438
- Ambros V. The functions of animal microRNAs. Nature 2004; 431:350 - 5; http://dx.doi.org/10.1038/nature02871; PMID: 15372042
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol 2007; 23:175 - 205; http://dx.doi.org/10.1146/annurev.cellbio.23.090506.123406; PMID: 17506695
- Zhang L, Hou DX, Chen X, Li DH, Zhu LY, Zhang YJ, et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA (vol 22, pg 273, 2012). Cell Res 2012; 22:273 - 4; http://dx.doi.org/10.1038/cr.2011.174
- LaMonte G, Philip N, Reardon J, Lacsina JR, Majoros W, Chapman L, et al. Translocation of sickle cell erythrocyte microRNAs into Plasmodium falciparum inhibits parasite translation and contributes to malaria resistance. Cell Host Microbe 2012; 12:187 - 99; http://dx.doi.org/10.1016/j.chom.2012.06.007; PMID: 22901539
- Sutton RE, Boothroyd JC. Evidence for trans splicing in trypanosomes. Cell 1986; 47:527 - 35; http://dx.doi.org/10.1016/0092-8674(86)90617-3; PMID: 3022935
- Pfeffer S, Zavolan M, Grässer FA, Chien M, Russo JJ, Ju J, et al. Identification of virus-encoded microRNAs. Science 2004; 304:734 - 6; http://dx.doi.org/10.1126/science.1096781; PMID: 15118162
- Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR. Kaposi’s sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci USA 2005; 102:5570 - 5; http://dx.doi.org/10.1073/pnas.0408192102; PMID: 15800047
- Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grässer FA, et al. Identification of microRNAs of the herpesvirus family. Nat Methods 2005; 2:269 - 76; http://dx.doi.org/10.1038/nmeth746; PMID: 15782219
- Grey F, Meyers H, White EA, Spector DH, Nelson J. A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS Pathog 2007; 3:e163; http://dx.doi.org/10.1371/journal.ppat.0030163; PMID: 17983268
- Murphy E, Vanícek J, Robins H, Shenk T, Levine AJ. Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: implications for latency. Proc Natl Acad Sci USA 2008; 105:5453 - 8; http://dx.doi.org/10.1073/pnas.0711910105; PMID: 18378902
- Choy EY, Siu KL, Kok KH, Lung RW, Tsang CM, To KF, et al. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J Exp Med 2008; 205:2551 - 60; http://dx.doi.org/10.1084/jem.20072581; PMID: 18838543
- Hansen A, Henderson S, Lagos D, Nikitenko L, Coulter E, Roberts S, et al. KSHV-encoded miRNAs target MAF to induce endothelial cell reprogramming. Genes Dev 2010; 24:195 - 205; http://dx.doi.org/10.1101/gad.553410; PMID: 20080955
- Nachmani D, Stern-Ginossar N, Sarid R, Mandelboim O. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe 2009; 5:376 - 85; http://dx.doi.org/10.1016/j.chom.2009.03.003; PMID: 19380116
- Samols MA, Skalsky RL, Maldonado AM, Riva A, Lopez MC, Baker HV, et al. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog 2007; 3:e65; http://dx.doi.org/10.1371/journal.ppat.0030065; PMID: 17500590
- Stern-Ginossar N, Elefant N, Zimmermann A, Wolf DG, Saleh N, Biton M, et al. Host immune system gene targeting by a viral miRNA. Science 2007; 317:376 - 81; http://dx.doi.org/10.1126/science.1140956; PMID: 17641203
- Xia T, O’Hara A, Araujo I, Barreto J, Carvalho E, Sapucaia JB, et al. EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res 2008; 68:1436 - 42; http://dx.doi.org/10.1158/0008-5472.CAN-07-5126; PMID: 18316607
- Gottwein E, Mukherjee N, Sachse C, Frenzel C, Majoros WH, Chi JTA, et al. A viral microRNA functions as an orthologue of cellular miR-155. Nature 2007; 450:1096 - 9; http://dx.doi.org/10.1038/nature05992; PMID: 18075594
- Skalsky RL, Samols MA, Plaisance KB, Boss IW, Riva A, Lopez MC, et al. Kaposi’s sarcoma-associated herpesvirus encodes an ortholog of miR-155. J Virol 2007; 81:12836 - 45; http://dx.doi.org/10.1128/JVI.01804-07; PMID: 17881434
- Otsuka M, Jing Q, Georgel P, New L, Chen JM, Mols J, et al. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity 2007; 27:123 - 34; http://dx.doi.org/10.1016/j.immuni.2007.05.014; PMID: 17613256
- Huang JL, Wang FX, Argyris E, Chen KY, Liang ZH, Tian H, et al. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat Med 2007; 13:1241 - 7; http://dx.doi.org/10.1038/nm1639; PMID: 17906637
- Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, et al. A cellular microRNA mediates antiviral defense in human cells. Science 2005; 308:557 - 60; http://dx.doi.org/10.1126/science.1108784; PMID: 15845854
- Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 2005; 309:1577 - 81; http://dx.doi.org/10.1126/science.1113329; PMID: 16141076
- Xue XY, Zhang QF, Huang YF, Feng L, Pan WQ. No miRNA were found in Plasmodiuml and the ones identified in erythrocytes could not be correlated with infection. Malar J 2008; 5:606 - 9