Abstract
Ribonuclease P RNA requires a sharply kinked RNA helix to make a loop-receptor interaction that creates the binding site for the substrate. In some forms of the ribozyme, this is accomplished by a k-turn, while others have a different element called the pk-turn. The structure of the pk-turn in RNase P of Thermotoga maritima is globally very similar to a k-turn, but lacks all the standard features of that structure, including long-range hydrogen bonds between the two helical arms. We show here that in an isolated RNA duplex, the pk-turn fails to adopt a tightly kinked structure, but rather is a flexible element. This suggests that the tertiary contacts of RNase P assist its folding into the required kinked structure. We find that we can replace the k-turn of the SAM-I riboswitch with the pk-turn, such that the resulting RNA retains its ability to bind SAM, although with lower affinity. We also find that we can replace the pk-turn of T. maritima RNase P with a standard k-turn (in either orientation) with retention of ribozyme activity. Thus, although the pk-turn cannot intrinsically fold into the kinked structure, it can be induced to fold correctly in context. And the pk-turn and k-turns can substitute functionally for one another.
Introduction
The complexity of RNA structure can be rationalized at one level by considering the ensemble as a set of helical sections connected by junctions,Citation1-Citation7 which are then locked together by tertiary interactions. If we define a junction as two or more helical sections connected by the continuity of individual strands, then the simplest of these is a two-way junction, in which there is a significant change in trajectory of the helix axis in a duplex, thereby introducing a marked bend or kink. Axial kinking facilitates long-range tertiary interactions that are essential to the overall architecture of the RNA, and can create sites for protein interaction and binding pockets for small molecules.
The kink turn (k-turn) is a very widespread motif that introduces a tight kink with an included angle of 50–60° in duplex RNA. There are multiple examples in rRNACitation8 and they are key elements in snoRNA species, U4 snRNACitation9 and several riboswitches.Citation10-Citation12 In the SAM-I riboswitch a k-turn introduces a kink into a long helix that allows its terminal loop to interact with a receptor and, thus, create the binding pocket for its ligand,Citation10 and disruption of the k-turn prevents its binding.Citation13 Other sequence elements have been identified in RNA structures that introduce sharp kinks, including the reverse kink-turnCitation14,Citation15 and most recently the pk-turn.Citation16 These can introduce kinks of comparable magnitude to the k-turn, but their local structure is quite different. Some may not be intrinsically kinked, but rather might be inherently flexible so that they can adopt a kinked geometry if stabilized by additional interactions in the RNA.
The standard k-turn comprises a typically 3-nucleotide bulge followed by G•A and A•G pairs on its 3′ side (). The G•A pairs are both trans sugar-edge:Hoogsteen pairs oriented to direct the minor groove edges of the adenine nucleobases into the minor groove of the opposing helix. Within the core of the structure there are a number of critical cross-strand hydrogen bonds involving 2'-hydroxyl groups bridging the interface.Citation17,Citation18 In free solution in the absence of metal ions the k-turn adopts an extended structure. However, the kinked structure can form stably in the presence of metal ions,Citation19,Citation20 bound proteinCitation21 or tertiary interactions.Citation13
Figure 1. Sequences of pk and k-turn motifs in RNA. (A) The sequences of the T. tengcongensis SAM-I riboswitch k-turn and the T. maritima RNase P pk-turn. The standard nucleotide nomenclature is shown for the k-turn,Citation17 while the nucleotide numbering in the pk-turn follows that used in.Citation16 (B) A scheme showing the secondary structure of the pk-turn in RNase P. The arrows are deduced hydrogen bonds. Note that there are no hydrogen bonds between the strands. (C) Parallel-eye stereoscopic views of the individual structures of the two strands of the pk-turn taken from the crystal structure of T. maritima RNase P (PDB code 3Q1Q).Citation22 Deduced hydrogen bonds are indicated by gray, broken lines. (D) Superposition of the structures of the T. maritima RNase P pk-turn (loops cyan, basepairs green) (PDB code 3Q1Q) and the T. tengcongensis SAM-I riboswitch k-turn (loop magenta, basepairs gray) (PDB entry 3GX5).
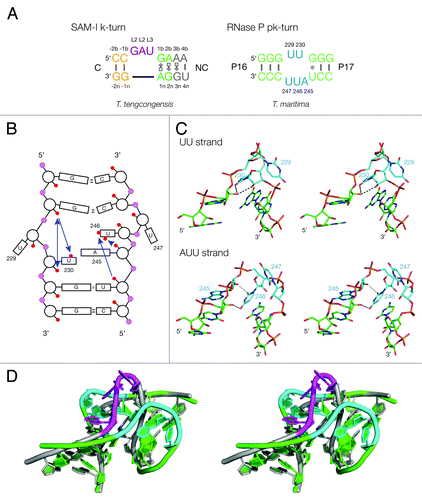
The structure of the pk-turn has been determined in the context of the complete RNase P holoenzyme of Thermotoga maritima;Citation22 it is an asymmetric loop that adopts a kink in the P16/P17 helix (). Rather analogously to the SAM-I riboswitch structure,Citation10 this allows the terminal loop of the P17 helix to dock into a receptor forming a long-range tertiary interaction that is required to form the correct architecture of the ribozyme.Citation16 This is likely to be critical to the ribozyme activity because the substrate binds at the adjacent loop L15. The sequence of the pk-turn is quite different from a k-turn, yet globally it has been shown that the structure can be superimposed with a k-turn with an RMSD of 1.3 Å. Indeed, analysis of many RNase P sequences indicates that some have a k-turn in place of the pk element, while some replace it with three- or four-way helical junctions.Citation16 This suggests that the pk and k-turns are functionally exchangeable. Yet the two structures differ fundamentally. The pk-turn is formally an HS3HS2 junction,Citation23 lacking G•A pairs and with unpaired loops that are very uridine-rich. In the T. maritima structure, the turns are made within the single-stranded loops essentially at a single nucleotide for both strands, in each case stacking a uridine nucleobase on the end of the helix. The only chemically feasible hydrogen bonds in the structure are relatively short-range interactions within the same strand (). For example, there is a bond between the O2' atoms of U229 and G231 on the U2 strand of length 2.8 Å and good geometry. Although the resolution of this crystal structure was not high, we can state with confidence that there are no long-range or inter-strand hydrogen bonds connecting the two helical arms, nor any possibility of A-minor interactions. The absence of such contacts bridging the helices suggests that the kinked geometry is unlikely to be stable as a free structure. More likely it is a point of flexibility that can be stabilized in a kinked conformation by further tertiary contacts.
We have therefore set out to explore the properties of the pk-turn experimentally. We find that it is unable to fold into a tightly kinked structure in isolation as a simple duplex upon addition of metal ions. However, we show that the pk-turn and k-turn may functionally substitute for each other in the SAM-I riboswitch and RNase P.
Results
The pk-turn element does not adopt a stable kinked conformation in isolation
The pk-turn adopts a strongly-kinked structure that is globally similar to a k-turn within the complete RNase P RNA (). This raises the question of whether this element can be induced to fold into the kinked conformation in isolation upon addition of metal ions (as can simple k-turns). Alternatively, it could be a point of flexibility that can be stabilized in the kinked form by tertiary interactions. A precedent for the latter behavior is set by the SAM G2nA k-turn, which is unable to fold in isolation but induced to adopt the kinked structure by tertiary interaction in situ in the riboswitch.Citation13 We therefore studied the pk-turn as an isolated duplex, using bulk-phase steady-state fluorescence resonance energy transfer (FRET), to see if it would adopt a kinked conformation on addition of metal ions.
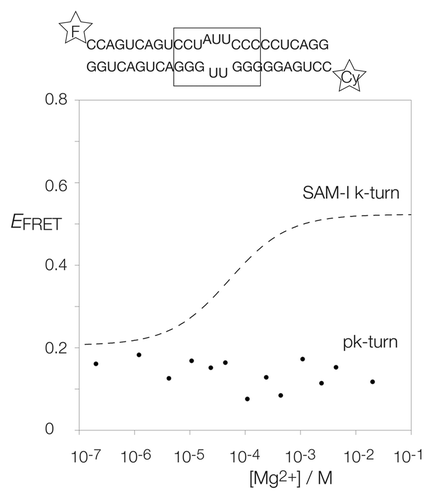
For this purpose, the pk-turn sequence was incorporated into the center of a short RNA double helix labeled with fluorescein (fluorescent donor) and Cy3 (acceptor) at the two 5′-termini. The FRET efficiency was then measured as a function of Mg2+ concentration (). Folding into a kinked geometry shortens the inter-fluorophore distance, leading to an increase in FRET efficiency (EFRET). The SAM-I k-turn undergoes an increase in EFRET as the concentration of Mg2+ is increased typical of many k-turns, showing the stabilization of the kinked geometry of the k-turn (broken line in ). In marked contrast, however, the pk-turn exhibits a low value of EFRET that does not increase with addition of Mg2+ (points in ) We conclude that in isolation, the pk-turn fails to adopt a kinked conformation that is stabilized by Mg2+ ions and, thus, it seems unable intrinsically to adopt the tightly kinked conformation. But since it has been shown to have a structure that is globally similar to a k-turn when part of the complete ribozyme, it is more likely to be a point of flexibility that can be stabilized in the kinked form by other influences such as tertiary interactions.
Figure 2. Analysis of ion-induced structural change in duplex RNA-containing a pk-turn studied by fluorescence resonance energy transfer in the steady-state. The pk-turn was centrally located (boxed in the sequence shown) within a 22 bp duplex of RNA terminally 5′-labeled with fluorescein (F, donor) and Cy3 (Cy, acceptor) fluorophores. FRET efficiency was measured in 90 mM Tris.borate (pH 8.3) with addition of MgCl2 to the indicated concentrations. The data are plotted as FRET efficiency (EFRET) as a function of Mg2+ concentration. Data for the SAM-I riboswitch k-turn are plotted (broken line) as the best fit to a two-state ion-induced transition, taken from.Citation13 Note that there is no increase in EFRET for the pk-turn-containing RNA at any Mg2+ concentration.
The pk-turn adopts a broad distribution of extended geometries that is largely unaffected by the presence of metal ions
We have used time-resolved fluorescence measurements of the same donor-acceptor-labeled pk-turn RNA to gain some insight into the distribution of conformations that are present as a function of solution conditions. We have compared the results with comparable measurements on the standard k-turn Kt-7 of H. marismortui. The decay of fluorescein fluorescence following a rapid excitation pulse was fitted to obtain distributions of excited state lifetimes (). Kt-7 in the absence of Mg2+ ions exhibits a distribution that comprises a single peak, with a mean lifetime of 3.3 ns. This corresponds to EFRET = 0.21, i.e., the extended, unfolded structure expected for the k-turn in these conditions. Upon addition of 2 mM Mg2+ to the sample, the distribution becomes bimodal, with a narrowed peak of 3.3 ns, and a second distribution with a mean lifetime of 0.95 ns. The latter corresponds to EFRET = 0.77, in good agreement with single-molecule analysis of the folded conformation of the k-turn.Citation24 Evidently, under these conditions, the solution contains similar fractions of folded and unfolded k-turn molecules.
Figure 3. Analysis of the distribution of fluorescein fluorescent lifetime distributions for pk-turn and k-turn-containing RNA as a function of the presence or absence of Mg2+ ions. The same fluorescein, Cy3 pk-turn-containing RNA and an equivalent species containing the H. marismortui Kt-7 were used in these experiments.
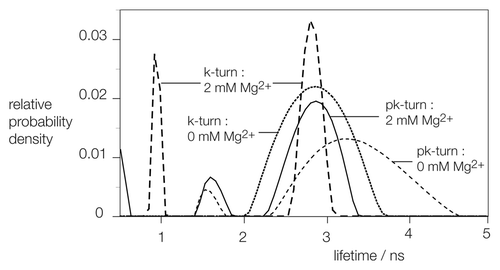
In the absence of Mg2+ ions, the pk-turn leads to a unimodal distribution of lifetimes, with a mean value of 3.7 ns. This is again consistent with an essentially extended conformation, and the distribution is a little wider than that for the k-turn under the same conditions. Addition of Mg2+ ions results in some narrowing of the distribution, but in contrast to the k-turn, there is no distribution of lifetime in the 1 ns region. Thus, the time-resolved FRET data provide no evidence for a fraction of tightly kinked RNA for the pk-turn, irrespective of the presence or absence of metal ions.
The pk-turn can functionally substitute for the k-turn of the SAM-I riboswitch
The SAM-I riboswitch contains a k-turn that is required to kink the P2 helix so that it can make a loop-receptor tertiary interaction.Citation10 If the k-turn is disrupted by a A1nC substitution, the riboswitch is no longer able to bind its SAM ligand as demonstrated by isothermal titration calorimetry (ITC).Citation13 Using molecular modeling, we examined the stereochemical feasibility of replacing the riboswitch k-turn with the T. maritima pk-turn sequence. The two structures can be well-superimposed (). Moreover, using computer-based molecular graphics we have constructed a model of the riboswitch in which the pk-turn replaces the natural k-turn with the helical trajectories preserved. We have therefore probed the ability of the pk-turn to substitute for the k-turn of the riboswitch experimentally, testing for the ability of the modified RNA to bind SAM.
We substituted the k-turn for the pk-turn in the Thermoanaerobacter tengcongensis SAM-I riboswitch by site-directed mutagenesis of the template plasmid, and transcribed the modified riboswitch RNA. ITC study shows that the pk-substituted riboswitch binds SAM in an exothermic reaction (), with a lowered affinity of 300 µM (). Thus, the pk-turn can clearly substitute for the k-turn with retention of activity, suggesting that it can accommodate the kinked structure required for the folding of the riboswitch. However, the binding affinity is 1,000-fold lower than for the natural riboswitch.
Figure 4. Replacement of the SAM-I riboswitch k-turn with the pk-turn. (A) A scheme shows the secondary structure and tertiary interaction of the SAM-I riboswitch, with the SAM-binding site indicated. The natural k-turn has been replaced by the pk turn in this structure. (B) Plots showing the results of microcalorimetric analysis of ligand binding to the SAM-I riboswitch containing the pk-turn. The upper panel shows the raw data for sequential injections of 2 µl volumes of a 5 µM solution of SAM into a 1.4 ml volume of 96 µM RNA solution in 50 mM HEPES (pH 7.5), 100 mM KCl, 10 mM MgCl2. This represents the differential of the total heat evolved for each SAM concentration (i.e., ∆H°). The lower panel presents the integrated heat data fitted to a single-site binding model (EquationEq. 1).
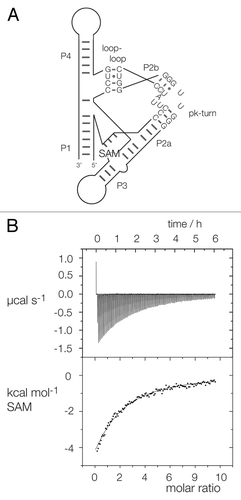
Table 1. Thermodynamic parameters for the SAM-I riboswitch
The k-turn can functionally substitute for the pk-turn in RNase P
Given that the pk-turn can substitute for the k-turn of the SAM-I riboswitch to some degree, we decided to explore the reverse substitution to see if the pk-turn of T. maritima RNase P could be replaced by a k-turn with retention of ribozyme activity. We superimposed the k-turn of the SAM-I riboswitch onto the pk-turn in the RNase P crystal structure in the two possible orientations by replacing the GGGUUGGG strand of the pk-turn with the bulged strand of the k-turn (termed kt-f) and by replacing the GGGUUGGG strand with the non-bulged strand of the k-turn (termed kt-r) (). The resulting models suggested that a k-turn could potentially substitute for the pk-turn in both orientations, allowing the required tertiary interaction to form without steric clash.
Figure 5. Ribozyme activity on RNase P in which the pk-turn has been replaced by a k-turn sequence in two orientations. (A) The secondary structure of the P15-P16-P17 section of RNase P showing the location of the pk-turn, and the tertiary interaction made by the terminal loop. The gray bars indicate basepairing, but the number of them is not significant. The ribozyme substrate binds at the loop L15. The sequence of the pk-turn and the forward and reverse oriented k-turns are shown. These are each located in RNase P in the same sense, i.e., P17 and its terminal loop is on the right-hand side as written. (B) Ribozyme activity of the natural and substituted forms of RNase P. Radioactive substrate (133 nt) and products (42 and 91 nt) were separated by gel electrophoresis following incubation with RNase P for 1 h. at 50°C. Substrate was loaded directly onto the gel without incubation (U), or after 1 h of incubation under assay conditions without added RNase P (S). The remaining samples were incubated for 1 h with RNase P, of native sequence including the pk-turn (pk), or with the pk-turn replaced by the SAM-I k-turn in the forward (kt-f) or reverse (kt-r) direction. Note that substrate is cleaved by all three ribozymes, at a slightly reduced level for the k-turn-containing forms. (C) Reaction progress plotted for the natural and substituted forms of RNase P. Symbols: natural ribozyme (containing the pk turn), closed circles; ribozyme containing the kt-f k-turn, closed squares; ribozyme containing the kt-r k-turn, open squares. The data are fitted to one or two exponential functions (lines).
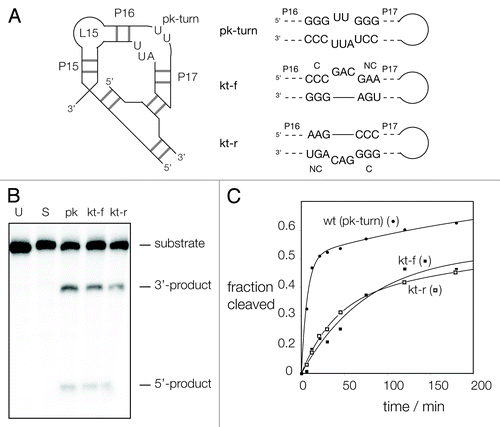
Ribozyme activity of the modified species was tested by analyzing the cleavage of a synthetic radioactively (α-32P)-labeled substrate RNA in the absence of the protein subunit in presence of 200 mM Mg2+ ions at 50°C, separating the products by PAGE (). Under the conditions of our assay, the unmodified ribozyme cleaved ~50% of the substrate at a rate of kobs = 0.11 ± 0.03 min−1. The data were fitted to two exponential functions (); we attribute a slow second rate of kobs = 0.004 min−1 to autolytic cleavage of the substrate under the assay conditions. The ribozymes with the pk-turn substituted by a k-turn were clearly active in either orientation. The substrate was cleaved to ~35%, with rates of kobs = 0.016 ± 0.003 min−1 and 0.023 ± 0.005 min−1 for the kt-f and kt-r variants of the ribozyme, respectively. Thus, the k-turn can functionally substitute for the pk-turn in RNase P.
Discussion
The pk-turn of T. maritima RNase P can form a closely similar kinked conformation to the k-turn in global terms ().Citation16 Yet, we show here that in isolation, this sequence element acts as a flexible point rather than adopting a fixed kink structure like the k-turn in the presence of metal ions, consistent with the lack of long-range hydrogen bonds between the two helical regions. Although there is no evidence for a significant population of a kinked form of the RNA as a simple duplex, evidently, tertiary contacts within the ribozyme can stabilize a structure that is strikingly similar to the k-turn. We have recently shown that formation of a k-turn structure on binding of L7Ae protein most likely occurs by conformational selection;Citation24 it is probable that this is also true for the pk-turn. We would expect that its inherent flexibility allows P17 to explore the surrounding environment and when its terminal loop encounters the receptor, it forms a persistent interaction and, thus, the k-turn-like conformation of the pk-turn is then stabilized. The folding of a G2nA variant of the SAM-I k-turn in the context of the riboswitch sets a precedent for the stabilization of local conformation by tertiary interactions in RNA.Citation13 Clearly, the pk-turn forms the required structure less easily than a k-turn. The 1,000-fold weaker binding of the SAM ligand to the pk-turn-containing riboswitch indicates a free energetic penalty of folding the pk-turn compared with its native k-turn of ~17 kJ mol−1.
We have shown that pk-turns and k-turns can functionally substitute for each other. SAM-I riboswitch with its k-turn replaced by the pk-turn can bind SAM, while RNase P with its pk-turn replaced by a k-turn is active. Remarkably, the k-turn can be inserted in either orientation into the ribozyme with very similar rates of substrate cleavage. It might be anticipated that the C-helix would be more likely to position the terminal loop better for contact with its receptor, but this is clearly not essential. Nevertheless, all the natural cases of RNase P with k-turns that we have examined have been oriented with this sense.
Although the pk-turn of T. maritima RNase P folds more weakly than k-turns, it is conceivable that in vivo it could be stabilized by the binding of an as-yet-unidentified protein. k-turns very frequently act as protein-binding sites, and it has been shown that RNase P of the archaeon Methanococcus maripaludis copurifies with the well-known k-turn binding protein L7Ae, and that addition of the protein to in vitro reactions greatly increase ribozyme activity.Citation25 An analogous interaction might occur for the pk-turn.
The lysine riboswitch may provide another example of structural diversity in kinking elements. Lafontaine and coworkersCitation11 have noted that in 32 riboswitches, there is a probable k-turn-forming sequence in the aptamer domain within helix P2 the terminal loop that makes an extensive loop-loop interaction with that of helix P3. They showed that the putative k-turn of Bacillus subtilus formed a kinked structure as an isolated RNA duplex, and could bind L7Ae protein. Yet, interestingly, when the structure of the lysine riboswitch of T. maritima was determined,Citation26,Citation27 the k-turn was not present, but its structural function was taken by a different strongly kinked element. The structure lacks G•A pairs, and has few long-range hydrogen bonds stabilizing the turn and better resembles a truncated three-way junction, so is perhaps unlikely to adopt a stable kinked conformation in isolation.
Taken together, our results suggest that there is more than one solution to the problem of how to introduce a sharp bend or kink in a helix. The k-turn can be an intrinsically kinked structure with a stable fold, although it is likely that k-turns that depart more widely from the consensus sequence require additional stabilization. On the other hand, their structural role can be filled by more flexible elements that achieve the same conformational purpose, and in doing so, become stabilized in the kinked form. The evolutionary choice between the different elements may be governed by the folding pathway for the various RNA species, and perhaps a varying role of protein stabilization.
Materials and Methods
Synthesis and purification of RNA
RNA for fluorescence spectroscopy was synthesized using t-BDMS phosphoramidite chemistry,Citation28 and subsequently deprotected and purified as described in.Citation29 RNA duplexes were hybridized by slow-cooling in 90 mM Tris.borate (pH 8.3), 1 mM EDTA (TBE buffer) and purified by non-denaturing gel electrophoresis using 20% 19:1 polyacrylamide in TBE buffer.
SAM-I RNA, and RNase P RNA were transcribed in vitro using T7 RNA polymerase from DNA templates amplified by PCR from plasmids bearing the required sequences generated synthetically. Transcribed RNA was purified by PAGE under denaturing conditions. Gels contained 19:1 polyacrylamide in the presence of TBE, 7 M urea. Polyacrylamide concentrations were 12% (w/v) for SAM-I riboswitch and 8% (w/v) for RNase P.
The substrate pTS-L(U-1) sequence was taken fromCitation30 (all sequences written 5′‒3′): GCUUCCCGAUAAGGGAGCAGGCCAGUAAAAAGCAUUACCCCAUGGAGUGAUUCCCGAGCGGCCAAAGGGAGCAGACUCUAAAUCUGCCGGUCAUCGACCUUCGAAGGUUCGAAUCCUUCUCUCUCCGCCACUU.
It was transcribed using the same method, but internally radioactively labeled with (α-32P)-GTP. The substrate was purified by denaturing PAGE in 12% (w/v) acrylamide.
Steady-state fluorescence spectroscopy
Fluorescence emission spectra were recorded from 0.5 ml of a 20 nM solution of fluorescein, Cy3-labeled RNA in 90 mM Tris.borate (pH 8.3) at 4°C as a function of MgCl2 concentration using an SLM Aminco 8100 spectrometer. Spectra were corrected for lamp fluctuations and instrumental variations, and polarization artifacts were avoided by setting excitation and emission polarizers crossed at 54.7°. Values of FRET efficiency were measured using the acceptor normalization methodCitation31 implemented in MATLAB.
The oligonucleotides used to construct the pk-turn-containing duplex were as follows:
Fluorescein-CCAGUCAGUCCUAUUCCCCCUCAGG and Cy3-CCUGAGGGGGUUGGGACUGACUGG.
Time-resolved fluorescence spectroscopy
Measurements of fluorescent lifetime were made on 200 nM solutions of RNA in 90 mM Tris.borate (pH 8.3) in the presence or absence of 2 mM MgCl2 at room temperature using a LifeSpex fluorimeter (Edinburgh Instruments). Samples were excited with a pulsed laser at 473 nm at a repetition frequency of 10 MHz. Data were acquired at 515 nm in 4,096 channels covering a time interval between 0‒50 ns after each pulse, until the peak intensity reached 100,000 counts. For each data set, the fluorescent decay was de-convoluted from the instrument response function and subjected to a lifetime distribution analysis using the FAST software (Edinburgh Instruments).
Calorimetry
Microcalorimetric measurement of SAM binding to the SAM-I riboswitch and variants were performed by isothermal titration calorimetry as described in.Citation13 The complete sequence of the riboswitch containing the pk-turn was:
GGCTTATCAAGAGAGGTCCTATTCCCACTGGGGGTTGGGACCCGGCAACCAGAAATGGTGCCAAUUCCUGCAGCGGAAACGUUGAAAGAUGAGCCG.
SAM was titrated against 0.96 mM riboswitch RNA concentration in 50 mM HEPES (pH 7.5), 100 mM KCl, 10 mM MgCl2 at 30°C. Data were fitted to a two-state binding model using MICROCAL Origin software. Individual heat changes ∆Q at constant pressure are given by:
(1)
∆Q = v. ΔH.(RNA).{[Ka. (SAM)in / 1+ Ka. (SAM)in] - [Ka. (SAM)ni-1 / 1+ Ka. (SAM)ni-1]}
where ΔH is the change in enthalpy, v is the reaction volume, Ka is the association constant for SAM binding and (SAM)i is the SAM concentration at the i th injection. The Hill coefficient n was fixed to unity.
Measurement of RNase P activity
The sequences of the modified RNase P ribozymes were:
RNase P kt-for
GGAGAGGAGCAGGCGGTCGCGGGGGCGCACACCTGCGCTTCCCGAGGAAAGTCCGGACTCTGGAGCGGGGTGCCGGGTAACGCCCGGGAGGGGTGACCCTCGGACAGGGCCATAGAGAAGAAGACCGCCCGGGGGGAAACTTCCGGGCAAGGGTGGAACGGTGGGGTAAGAGCCCACCAGCGTCGGGGCAACCCGGCGGCTTGGCAACCCCCACCTGGAGCAAGGCCAAGCAGGCCCGACGAATCGCTCCCGGAGGGCCGGGTTGGCCGCTTGAGGTGTGCGGTAACGCACACCCCAGATTGATGACCGCCCACGACAGAATCCGGCTTATGCTCCTCTCCCGTG and, RNase P kt-rev GGAGAGGAGCAGGCGGTCGCGGGGGCGCACACCTGCGCTTCCCGAGGAAAGTCCGGACTCTGGAGCGGGGTGCCGGGTAACGCCCGGGAGGGGTGACCCTCGGACAGGGCCATAGAGAAGAAGACCGCCCGGGGGGAAACTTCCGGGCAAGGGTGGAACGGTGGGGTAAGAGCCCACCAGCGTCGGGGCAACCCGGCGGCTTGGCAACCCCCACCTGGAGCAAGGCCAAGCAGGGAGGGTCGCTCCCCCCGACGAACCGGGTTGGCCGCTTGAGGTGTGCGGTAACGCACACCCCAGATTGATGACCGCCCACGACAGAATCCGGCTTATGCTCCTCTCCCGTG.
RNase P ribozyme activity was studied using a protocol slightly adapted from reference Citation32. 0.18 μM substrate and 1 μM ribozyme were individually incubated in 40 mM HEPES (pH 7.5, adjusted using KOH), 400 mM NH4Cl (RX buffer) at 65°C for 10 min. The reaction was initiated by mixing 36 nM substrate with 0.3 µM ribozyme in the presence of 200 mM MgCl2 in RX buffer at 50°C. Aliquots were removed at the specific times and the reaction terminated by addition of two volumes of 7 M urea, 15% (v/v) formamide, 100 mM EDTA. Substrate and product were separated by electrophoresis on 12% (w/v) acrylamide gels under denaturing conditions, and quantified by exposure to storage phosphor plates and phosphorimaging using a Typhoon 9600FLS. Intensities were corrected for background and the number of guanine nucleotides contained within the strands. Reaction progress curves were fitted to single or double exponential functions as required.
Molecular modeling
Molecular modeling of pk-turn and k-turn structures was performed using PyMol.Citation33
Acknowledgments
We thank Stephanie Schorr for discussion, Scott McPhee for RNA synthesis and Cancer Research UK, The Wellcome Trust and HFSP for financial support of our work.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Duckett DR, Murchie AIH, Lilley DMJ. The global folding of four-way helical junctions in RNA, including that in U1 snRNA. Cell 1995; 83:1027 - 36; http://dx.doi.org/10.1016/0092-8674(95)90218-X; PMID: 8521503
- Lilley DMJ. Structures of helical junctions in nucleic acids. Q Rev Biophys 2000; 33:109 - 59; http://dx.doi.org/10.1017/S0033583500003590; PMID: 11131562
- Lescoute A, Westhof E. Topology of three-way junctions in folded RNAs. RNA 2006; 12:83 - 93; http://dx.doi.org/10.1261/rna.2208106; PMID: 16373494
- Chu VB, Lipfert J, Bai Y, Pande VS, Doniach S, Herschlag D. Do conformational biases of simple helical junctions influence RNA folding stability and specificity?. RNA 2009; 15:2195 - 205; http://dx.doi.org/10.1261/rna.1747509; PMID: 19850914
- Laing C, Schlick T. Analysis of four-way junctions in RNA structures. J Mol Biol 2009; 390:547 - 59; http://dx.doi.org/10.1016/j.jmb.2009.04.084; PMID: 19445952
- Masquida B, Beckert B, Jossinet F. Exploring RNA structure by integrative molecular modelling. N Biotechnol 2010; 27:170 - 83; http://dx.doi.org/10.1016/j.nbt.2010.02.022; PMID: 20206310
- Ouellet J, Melcher SE, Iqbal A, Ding Y, Lilley DMJ. Structure of the three-way helical junction of the hepatitis C virus IRES element. RNA 2010; 16:1597 - 609; http://dx.doi.org/10.1261/rna.2158410; PMID: 20581129
- Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO J 2001; 20:4214 - 21; http://dx.doi.org/10.1093/emboj/20.15.4214; PMID: 11483524
- Vidovic I, Nottrott S, Hartmuth K, Lührmann R, Ficner R. Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Mol Cell 2000; 6:1331 - 42; http://dx.doi.org/10.1016/S1097-2765(00)00131-3; PMID: 11163207
- Montange RK, Batey RT. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature 2006; 441:1172 - 5; http://dx.doi.org/10.1038/nature04819; PMID: 16810258
- Blouin S, Lafontaine DA. A loop loop interaction and a K-turn motif located in the lysine aptamer domain are important for the riboswitch gene regulation control. RNA 2007; 13:1256 - 67; http://dx.doi.org/10.1261/rna.560307; PMID: 17585050
- Smith KD, Lipchock SV, Ames TD, Wang J, Breaker RR, Strobel SA. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat Struct Mol Biol 2009; 16:1218 - 23; http://dx.doi.org/10.1038/nsmb.1702; PMID: 19898477
- Schroeder KT, Daldrop P, Lilley DMJ. RNA tertiary interactions in a riboswitch stabilize the structure of a kink turn. Structure 2011; 19:1233 - 40; http://dx.doi.org/10.1016/j.str.2011.07.003; PMID: 21893284
- Strobel SA, Adams PL, Stahley MR, Wang J. RNA kink turns to the left and to the right. RNA 2004; 10:1852 - 4; http://dx.doi.org/10.1261/rna.7141504; PMID: 15547133
- Antonioli AH, Cochrane JC, Lipchock SV, Strobel SA. Plasticity of the RNA kink turn structural motif. RNA 2010; 16:762 - 8; http://dx.doi.org/10.1261/rna.1883810; PMID: 20145044
- Meyer M, Westhof E, Masquida B. A structural module in RNase P expands the variety of RNA kinks. RNA Biol 2012; 9:254 - 60; http://dx.doi.org/10.4161/rna.19434; PMID: 22336704
- Liu J, Lilley DMJ. The role of specific 2′-hydroxyl groups in the stabilization of the folded conformation of kink-turn RNA. RNA 2007; 13:200 - 10; http://dx.doi.org/10.1261/rna.285707; PMID: 17158708
- Daldrop P, Lilley DMJ. The plasticity of a structural motif in RNA: Structural polymorphism of a kink turn as a function of its environment. RNA 2013; 0; In the press PMID: 23325110
- Matsumura S, Ikawa Y, Inoue T. Biochemical characterization of the kink-turn RNA motif. Nucleic Acids Res 2003; 31:5544 - 51; http://dx.doi.org/10.1093/nar/gkg760; PMID: 14500816
- Goody TA, Melcher SE, Norman DG, Lilley DMJ. The kink-turn motif in RNA is dimorphic, and metal ion-dependent. RNA 2004; 10:254 - 64; http://dx.doi.org/10.1261/rna.5176604; PMID: 14730024
- Turner B, Melcher SE, Wilson TJ, Norman DG, Lilley DMJ. Induced fit of RNA on binding the L7Ae protein to the kink-turn motif. RNA 2005; 11:1192 - 200; http://dx.doi.org/10.1261/rna.2680605; PMID: 15987806
- Reiter NJ, Osterman A, Torres-Larios A, Swinger KK, Pan T, Mondragón A. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature 2010; 468:784 - 9; http://dx.doi.org/10.1038/nature09516; PMID: 21076397
- Lilley DMJ, Clegg RM, Diekmann S, Seeman NC, von Kitzing E, Hagerman P. Nomenclature Committee of the International Union of Biochemistry: A nomenclature of junctions and branchpoints in nucleic acids. Recommendations 1994. Eur J Biochem 1995; 230:1 - 2; http://dx.doi.org/10.1111/j.1432-1033.1995.tb20526.x; PMID: 7601087
- Wang J, Fessl T, Schroeder KT, Ouellet J, Liu Y, Freeman ADJ, et al. Single-molecule observation of the induction of k-turn RNA structure on binding L7Ae protein. Biophys J 2012; 103:2541 - 8; http://dx.doi.org/10.1016/j.bpj.2012.11.006; PMID: 23260056
- Cho IM, Lai LB, Susanti D, Mukhopadhyay B, Gopalan V. Ribosomal protein L7Ae is a subunit of archaeal RNase P. Proc Natl Acad Sci USA 2010; 107:14573 - 8; http://dx.doi.org/10.1073/pnas.1005556107; PMID: 20675586
- Garst AD, Héroux A, Rambo RP, Batey RT. Crystal structure of the lysine riboswitch regulatory mRNA element. J Biol Chem 2008; 283:22347 - 51; http://dx.doi.org/10.1074/jbc.C800120200; PMID: 18593706
- Serganov A, Huang L, Patel DJ. Structural insights into amino acid binding and gene control by a lysine riboswitch. Nature 2008; 455:1263 - 7; http://dx.doi.org/10.1038/nature07326; PMID: 18784651
- Beaucage SL, Caruthers MH. Deoxynucleoside phosphoramidites - a new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Lett 1981; 22:1859 - 62; http://dx.doi.org/10.1016/S0040-4039(01)90461-7
- Wilson TJ, Zhao Z-Y, Maxwell K, Kontogiannis L, Lilley DMJ. Importance of specific nucleotides in the folding of the natural form of the hairpin ribozyme. Biochemistry 2001; 40:2291 - 302; http://dx.doi.org/10.1021/bi002644p; PMID: 11329299
- Brännvall M, Kikovska E, Wu S, Kirsebom LA. Evidence for induced fit in bacterial RNase P RNA-mediated cleavage. J Mol Biol 2007; 372:1149 - 64; http://dx.doi.org/10.1016/j.jmb.2007.07.030; PMID: 17719605
- Clegg RM. Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol 1992; 211:353 - 88; http://dx.doi.org/10.1016/0076-6879(92)11020-J; PMID: 1406315
- Paul R, Lazarev D, Altman S. Characterization of RNase P from Thermotoga maritima.. Nucleic Acids Res 2001; 29:880 - 5; http://dx.doi.org/10.1093/nar/29.4.880; PMID: 11160919
- DeLano WL. The PyMOL Molecular Graphics System. Palo Alto, CA: DeLano Scientific, 2002.