Abstract
Recently, it has been shown that tRNA molecules can be processed into small RNAs that are derived from both the 5′ and 3′ termini. To date, the function of these tRNA fragments (tRFs) derived from the 5′ end of tRNA has not been investigated in depth. We present evidence that conserved residues in tRNA, present in all 5′ tRFs, can inhibit the process of protein translation without the need for complementary target sites in the mRNA. These results implicate 5′ tRFs in a new mechanism of gene regulation by small RNAs in human cells.
Introduction
In the past decade, many different classes of small RNA have been described, and in many cases they have emerged as important regulators of gene expression (reviewed in ref. Citation1). Small RNAs are typically 19–35 nt in length and three main classes have been studied in depth: microRNAs (miRNAs), endogenous small interfering RNAs (endo-siRNAs) and Piwi-interacting small RNAs (piRNAs). siRNAs and miRNAs are created by similar mechanisms, and in both these cases the final step is the action of the endonuclease Dicer.Citation2,Citation3 piRNAs have a separate biogenesis mechanism that only involves Argonaute proteins.Citation4 Once produced, these different types of small RNA are loaded into an Argonaute protein through which they exert their biological effect, typically repression of gene expression.Citation5-Citation7
Recently a number of new types of small RNA have been described, derived from longer cellular RNAs. These non-canonical sources include small nucleolar RNA,Citation8 rRNA and tRNA (tRNA) from yeast TRAMP mutantsCitation9 and tRNA in embryonic stem and transformed mammalian cells.Citation10-Citation18 Work is now being undertaken to understand how these new RNAs fit into the small RNA world. In many cases they are produced by the action of Dicer in a similar manner to siRNAs and miRNAs, suggesting they may have similar gene regulatory functions, and indeed some are incorporated into Argonaute proteins. However, other nucleases have also been implicated in the generation of these small RNA species.Citation19,Citation20
Three general classes of small tRNA fragment (tRF) have been described: those produced by RNase Z cleavage at the 3′ end of the pre-tRNA transcript (3′ U tRFs), those produced by cleavage of mature tRNA at the 3′ end (3′ CCA tRFs) and those produced by cleavage at the 5′ end (5′ tRFs) (reviewed in refs. Citation21 and Citation22). 5′ and 3′ tRFs have been reported to be produced by the action of DicerCitation12,Citation17,Citation20 and other nucleases, such as angiogenin,Citation19 and it is possible that a combination of different nucleases are involved in generating different molecules of these species. 3′ U and 3′ CCA tRFs have both been shown to associate with Argonautes, although with different Argonaute preferences to human miRNAs.Citation17 3′ CCA tRFs can silence a suitable complementary target,Citation23 whereas 3′ U tRFs preferentially associate with Ago3/4 and are believed to only silence a target if a complementary (“sense”) small RNA is transfected in order to direct them into Ago2.Citation17
In addition to short tRFs, tRNAs have been reported to be cleaved into half-tRNA molecules in stress conditions in a wide variety of eukaryotic cells.Citation19,Citation24,Citation25 These half-tRNAs are produced by the action of angiogenin near the anticodon loop, and the 5′ halves have been reported to specifically cause eIF2α-independent translation repression and stress granule assembly.Citation26,Citation27 Some of these 5′ tRNA halves have been described to bind eIF4G (a component of the cap-binding initiation factor complex eIF4F), and hence titrate it away from mRNA and reduce the rate of translation initation.Citation27 This effect is dependent on a oligo(G) tract of at least four residues at the 5′ end of the half-tRNA (and by extension, its parent tRNA). These studies indicate that tRNA-derived RNA molecules are, in some circumstances, used in the global control of gene expression.
No function has yet been found for 5′ tRFs, and we have previously shown that they are only weakly associated with Argonaute proteins.Citation12 Here we present data that shows that they do not function as classical siRNAs or miRNAs; however, they can inhibit the translation of reporter genes in vitro and in vivo, an effect that does not require any complementary target sites in the reporter sequence, but does require a universally conserved “GG” dinucleotide in the tRF. This opens the possibility that 5′ tRFs, like other tRNA-derived fragments, are involved in regulating gene expression.
Results
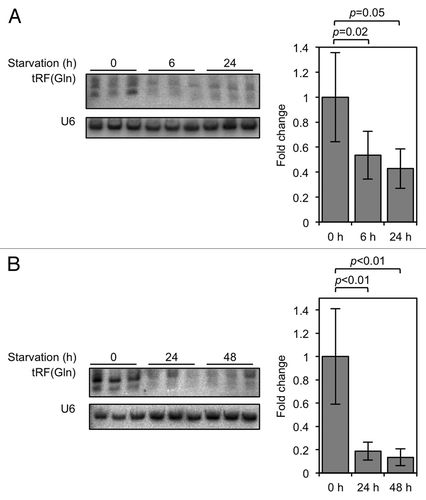
3′ U tRFs have previously been reported to be downregulated in cell culture if serum levels are decreased, and upregulated if the RNA polymerase III-specific transcription factor Brf1 is overexpressed.Citation14,Citation17 To find out if 5′ tRF levels are also responsive to conditions of cell stress, we completely starved cells of either L-glutamine or serum and assayed the levels of the 5′ tRF, tRF(Gln). [tRF(Gln) is 19 bases in length, and in previous workCitation12 we showed that tRF(Gln) is quite highly-expressed in HeLa cells. We therefore use it as an example of an endogenous 5′ tRF.] The results in show that tRF(Gln) levels decreased in both starvation conditions. The responsiveness of this 5′ tRF to conditions of varied cell proliferation rate is similar to previous results from 3′ U tRFs, and raises the possibility that different types of tRFs are regulated by similar biological stresses.Citation14,Citation17
Figure 1. tRF(Gln) expression is decreased in conditions of slowed cell proliferation. (A) HeLa cells were grown in DMEM lacking L-glutamine for the indicated times, and tRF(Gln) levels assayed by northern blot. (B) HeLa cells were grown in DMEM lacking serum for the indicated times, and tRF(Gln) levels assayed by northern blot. Quantifications of tRF(Gln) levels, normalized to the non-coding RNA U6, are shown adjacent to the blots. Error bars show standard deviations, and p values were calculated using Student's t-test (two-tailed).
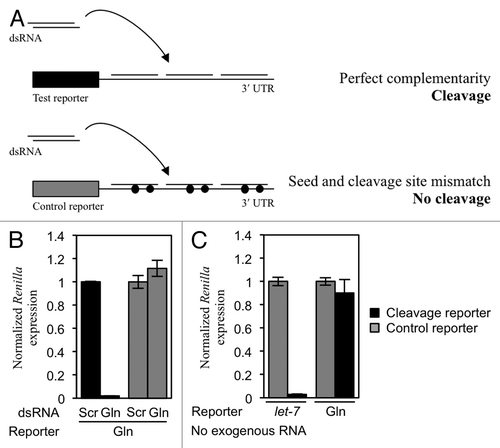
To investigate the function of 5′ tRFs, we started by testing whether they have a similar role to siRNAs or miRNAs in the regulation of gene expression. We have previously shown that 5′ tRFs associate with Argonaute proteins, albeit only very weakly.Citation12 In order to test if 5′ tRFs could inhibit gene expression in a manner similar to miRNAs, we made two reporter constructs. These constructs contain Renilla and firefly luciferase genes under the control of independent promoters, and have target sites for tRF(Gln) cloned into the 3′ UTR of the Renilla luciferase gene. One reporter had three perfectly complementary sites to tRF(Gln), and the other had six sites complementary to the putative seed region of the same tRF but containing a bulge outside of this region (Fig. S1). If 5′ tRFs act in a similar manner to miRNAs, the construct containing perfectly complementary sites would be targeted by RNAi, and the construct containing mismatched sites would undergo translational repression and degradation. We also created negative controls for both these constructs which contained mutations that would prevent both RNAi and translational repression.
First, the construct containing perfectly complementary sites (the siRNA reporter) was transfected into cells, and a double-stranded siRNA mimic of tRF(Gln) was cotransfected. We transfected this sequence as a double-stranded siRNA in order to force loading of the sequence into Argonautes.Citation28 In this experiment, the measured activity of Renilla luciferase was found to be sensitive to the siRNA (), indicating that the target sites were accessible and could target the mRNA for repression. We then transfected the reporter construct into cells without siRNA cotransfection, to measure the ability of endogenous tRF(Gln) to cause RNAi. When we did this, we observed no difference in relative expression between the reporter construct and the control (), indicating that endogenous tRF(Gln) does not efficiently induce RNAi.
Figure 2. tRF(Gln) does not cause siRNA-like cleavage of a reporter construct. (A) Experimental design. HeLa cells were transfected with a reporter plasmid containing three perfectly complementary target sites to tRF(Gln). When an dsRNA mimic of tRF(Gln) is co-transfected, the reporter is cleaved leading to a reduction in Renilla luciferase expression. This does not happen with a control reporter containing a mismatch in the seed sequence and at the cleavage site (represented as a filled circle). (B) The cleavage reporter construct was co-transfected with either a scrambled dsRNA (Scr), or an dsRNA mimic of tRF(Gln) (Gln). (C) Cleavage reporter constructs containing sites perfectly complementary to either the miRNA let-7 or tRF(Gln) were co-transfected into cells without cotransfection of dsRNA, in order to assay the activity of endogenous molecules. Separately, control reporters for let-7 and tRF(Gln) were transfected; these control reporters contain two mismatches in their respective target sites to render them insensitive to repression (gray bars). Results shown are the mean of three independent experiments, and error bars represent standard deviations.
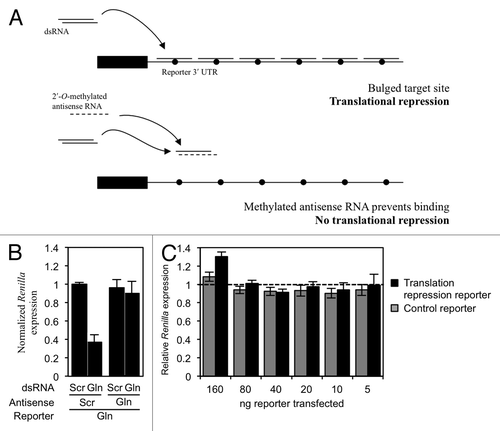
Next we tested if endogenous tRF(Gln) could regulate the expression of the reporter construct that contains six mismatched sites (the translation repression reporter). We took advantage of the fact that 2′-O-methylated oligonucleotides antisense to a small RNA inhibit the function of that small RNA.Citation29 We first validated the construct by co-transfecting it into HeLa cells with a dsRNA containing the tRF(Gln) sequence. In this experiment, the expression of Renilla luciferase was lower when we also transfected a scrambled antisense RNA than when we transfected one complementary to the tRF(Gln) sequence (). This showed that the plasmid that we designed is an appropriate reporter to test for miRNA-like action of tRF(Gln). We then performed this experiment in the absence of the exogenous dsRNA to test the ability of endogenous tRF(Gln) to inhibit translation. When this was done, there was no difference in normalized Renilla expression between the experiments with the scrambled and the anti-tRF(Gln) antisense RNAs [; a ratio of 1 indicates no tRF(Gln)-dependent translational repression]. This shows that endogenous tRF(Gln) does not regulate gene expression in a miRNA-like manner.
Figure 3. tRF(Gln) does not repress translation of a reporter construct with bulged target sites in the 3′ UTR. (A) Experimental design. HeLa cells were transfected with a reporter plasmid containing six bulged tRF(Gln) target sites, which is predicted to be sensitive to miRNA-like translational repression. In control experiments, a methylated antisense RNA was co-transfected to prevent small RNAs from binding the target sites. The bulge is represented as a filled circle. (B) The translation reporter was co-transfected into HeLa cells with either a scrambled dsRNA (Scr) or a mimic of tRF(Gln) (Gln), and also the indicated antisense RNA. (C) To assay the activity of endogenous molecules, the indicated amounts of a reporter plasmid containing six bulged miRNA-like sites for tRF(Gln) was transfected into HeLa cells (black bars). Renilla expression was assayed separately in the presence of a co-transfected scrambled antisense RNA, and with an antisense RNA containing the reverse complement of tRF(Gln). The relative Renilla expression was calculated as the normalized expression obtained with the scrambled antisense RNA, divided by that obtained with the Gln antisense RNA (see Materials and Methods), such that a value of 1 indicates no tRF(Gln)-dependent repression (dashed line). The same experiment was performed with a control reporter containing a seed mismatch (gray bars). Results shown are the mean of three independent experiments, and error bars represent standard deviations.
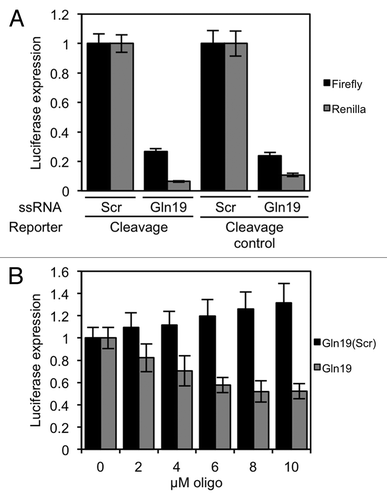
While carrying out these experiments, we noticed that when a 19-base synthetic RNA with the same sequence as tRF(Gln) (which we named Gln19 to distinguish the synthetic RNA from the endogenous molecule) was transfected into cells, the absolute activity of both firefly and Renilla luciferase in the lysate decreased. Importantly, the synthetic RNA had a repressive effect on these reporter genes regardless of whether the 3′ UTR contained engineered complementary target sites or not (). We also tested whether this RNA had a direct effect on protein translation in vitro using translation-competent HeLa cell extract,Citation30 and found that translation was indeed inhibited in this system by the RNA molecule but not by its scrambled sequence ().
Figure 4. Gln19 can repress translation of Renilla and firefly luciferases that do not contain complementary target sites. (A) HeLa cells were transfected with a reporter plasmid, and 19-base single-stranded RNA molecules containing either the tRF(Gln) sequence or a scrambled sequence. The firefly gene contained no target sites, and the Renilla gene contained either three perfect Gln target sites or mutated control versions of the same, as indicated. The expression of Renilla and firefly luciferases were measured separately. (B) In vitro translation reactions were performed with the indicated concentrations of Gln19 or Gln19(Scr) present in the reaction. The messenger did not contain any Gln19 target sites. Results shown are the mean of three independent experiments, and error bars represent standard deviations.
As we had not inserted target sites into these reporter genes that proved to be sensitive to Gln19, we decided to check that the repression was not due to RNA base-pairing between Gln19 and any cryptic target sites that may be present in the coding region of our reporters. To do this, we identified the sequence in Renilla that was most complementary to Gln19 using the RNAhybrid software.Citation31 The sequence complementarity between this “target site” and Gln19 is poor, but to ensure that repression was not taking place through this sequence, we introduced silent mutations in the reporter construct and assayed the ability of Gln19 to repress translation of mRNAs containing these mutations (Fig. S2A). Repression still occurred with the same efficacy under these conditions (Fig. S2B). This result indicates that the repressive effect of the Gln19 RNA molecule that we observed is a non-miRNA-like mode of action, and not due to recognition of near-complementary target sites in the mRNA. We also verified that the decrease in luciferase activity was not due to an effect of the synthetic RNA on the enzyme itself. To do this, we measured the effect of adding Gln19 to in vitro translated Renilla luciferase after translation was complete. In these conditions, the luciferase enzymatic activity was unperturbed (Fig. S2C).
Because the RNA sequence contained in the 5′ tRF, tRF(Gln), is able to repress translation, we investigated the specificity of the effect by seeing if other 5′ tRFs are able to similarly inhibit translation. To do this, we synthesized RNA molecules that corresponded to 5′ tRFs from different tRNA isotypes. We also checked that the translation repression was specific to 5′ tRF sequences, by synthesizing both an RNA that corresponds to the 19 bases in tRNAGln that starts after the bona fide tRF(Gln) sequence (Glnmid), and an RNA that corresponds to a reported 3′ U tRF (tRF-1001).Citation14,Citation17 The predicted structures of all RNA molecules used in these experiments are shown in Fig. S3. When these RNA molecules were tested in our translation assay, three out of the four bona fide 5′ tRFs inhibited translation (the exception being Glu19). Additionally, neither tRF-1001 nor Glnmid caused translation inhibition (), indicating that the translation repression effect was restricted to 5′ tRF sequences.
Figure 5. Translation repression requires a 'GG' dinucleotide that is conserved in tRFs. (A) In vitro translation reactions were performed in the presence of the indicated 19-mer oligonucleotides. (B) Diagrams of the Gln19-derived oligonucleotides that were used to find sequence requirements for translation repression. The mutated bases are highlighted. (C) In vitro translation reactions were performed with the Gln19-derived oligonucleotides. Gln19(mGG) showed a severe defect in its ability to repress translation. (D and E) In vitro translation reactions were performed with (D) Lys19 and (E) Val19, and their corresponding mutants. In both cases, translation repression required a 3′ “GG” dinucleotide. In all reactions, the concentrations of oligonucleotide used were 0, 2.5, 5 and 10 µM. Results shown are the mean of three independent experiments, and error bars represent standard deviations.
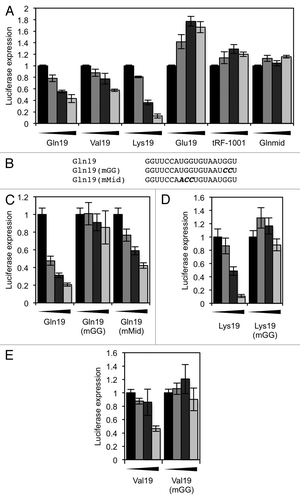
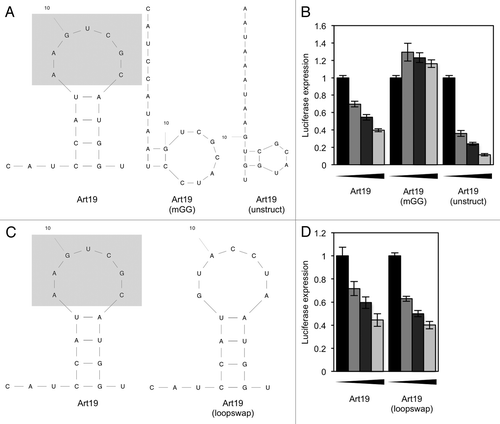
The fact that most of the 5′ tRFs we tested were able to repress translation suggests that the effect is conferred by conserved sequence or structural features. tRNAs contain a number of conserved residues that are required for their correct 3-dimensional folding and recognition by the translation machinery.Citation32 We synthesized RNA molecules that contained mutations of some conserved tRNA residues. All 5′ tRFs contain a “GG” dinucleotide sequence either at positions 17–18 or 18–19, depending on the parent tRNA; the molecule Gln(mGG) had these residues mutated. We also made Gln(mMid), which contained a mutation of UGG8-10, another conserved sequence. When we tested these synthetic RNAs in our translation assay, Gln(mGG) proved to be inactive in translation inhibition whereas Gln(mMid) could still repress translation, albeit less efficiently (). From this we concluded that the 3′ terminal “GG” dinucleotide appears necessary for translation inhibition by Gln19. To see if the 3′ terminal “GG” dinucleotide is necessary in other 5′ tRFs, we mutated the corresponding dinucleotide in Lys19 and Val19; this similarly abolished the ability of these molecules to repress translation (). Although the 3′ terminal “GG” dinucleotide is necessary for 5′ tRFs to be able to repress translation in all the instances we tested, it is not sufficient, as Glu19 does not repress translation despite containing a 3′ “GG.”
We next made a synthetic RNA that was predominantly not derived from tRNA sequence (although the loop sequence in the predicted structure was derived from Gln19) which contained a 3′ ‘GG’ dinucleotide (Art19). This was also able to repress translation and, in a similar manner to before, this effect was ablated in when the “GG” dinucleotide was mutated (). A 3′ “GG” dinucleotide is therefore able in some cases to make an otherwise inactive RNA molecule repress translation. RNA secondary structure does not appear important, as a mutation of Art19 with substantially disrupted predicted secondary structure [Art19(unstruct)] was still able to inhibit translation (). The RNA was also still able to inhibit translation when we replaced the Gln19-derived loop sequence to ensure that we had not kept any other recognition motifs [Art19(loopswap)] ().
Figure 6. Translation assays using a non-tRNA-derived sequence. (A) A 19-base oligonucleotide (Art19), containing a 3′ “GG,” was designed with only the loop sequence (gray box) derived from tRNA. Related oligos with the 3′ “GG” mutated, and an oligo containing the 3′ “GG” but designed to have less secondary structure, were also designed. (B) The oligonucleotides shown in (A) were used in an in vitro translation reaction. (C) An oligonucleotide was designed that is derived from Art19, but with the loop sequence changed to a non-tRNA-derived sequence. (D) The oligonucleotides shown in (C) were used in an in vitro translation reaction. In all reactions, the concentrations of oligonucleotide used were 0, 2.5, 5 and 10 µM. Results shown are the mean of three independent experiments, and error bars represent standard deviations.
In order to eliminate potential mechanisms by which 5′ tRFs cause translation repression, we looked to see if either the reporter gene mRNA, or the corresponding tRNA, is affected by the presence of 5′ tRFs. mRNA deadenylation and degradation plays a major contribution to the action of miRNAs (reviewed in ref. Citation33). To see a similar process occurs with 5′ tRFs, we assayed the level of Renilla mRNA by RT-qPCR at the end of an in vitro translation reaction that contained either Gln19 (to repress translation) or Gln19(mGG) (as a control). We used reverse transcription primers that produce cDNA either from total mRNA, or from polyadenylated mRNA. As can be seen in , there was no difference in either the total or polyadenylated mRNA levels between the samples after the translation reaction.
Figure 7. Translation repression by 5′ tRFs does not occur by mRNA or tRNA degradation, and does not involve the mRNA cap structure or poly(A) tail. (A) tRFs do not trigger mRNA deadenylation or degradation. An in vitro translation reaction was performed in the presence of 10 µM Gln19 or Gln19(mGG) and total and polyadenylated Renilla luciferase mRNA quantified by qPCR, using random hexamers or dT20 reverse transcription primers respectively. (B) tRFs do not change tRNA abundance. HeLa cells were transfected with either Gln19 or Gln19(Scr), and after 24 h, tRNA(Gln) quantified using qPCR are shown normalized to 5S abundance. (C) tRFs do not trigger a tRNA charging defect. An in vitro translation reaction was performed in the presence of either buffer, 10 µM Gln19 or 10 µM Gln19(mGG). The amounts of charged tRNA(Gln) and uncharged tRNA(Gln) in each sample were resolved using an acidic gel. (D) mRNAs were synthesized in vitro that contain either a cap analog plus a poly(A) tail, a cap analog and no poly(A) tail, or a non-functional cap analog and no poly(A) tail. These mRNAs were used in an in vitro translation reaction in the presence of Gln19. The concentrations of oligonucleotide used were 0, 2.5, 5 and 10 µM. Results shown are the mean of three independent experiments, and error bars represent standard deviations. Cap: m7G(5′)ppp(5′)G. ACap: A(5′)ppp(5′)G. p(A): poly(A) tail.
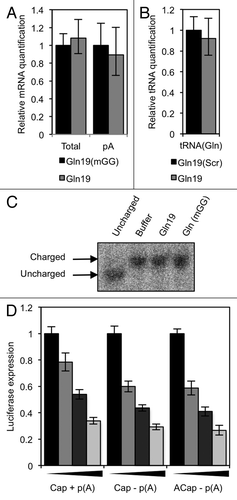
As 5′ tRFs are tRNA-derived, we then looked to see if transfection of 5′ tRFs has an effect on the corresponding mature tRNA level. Using a qPCR-based assay, we measured the levels of tRNAGln after transfecting cells with either Gln19 or Gln19(Scr). shows that the tRNA levels were the same in both samples. We also measured tRNAGln aminoacylation using an acid gel-based system,Citation34 to see if 5′ tRFs cause a tRNA charging defect, but saw no difference in tRNAGln charge state when Gln19 was added to the lysate ().
As the levels of these major translation substrates are unchanged by addition of 5′ tRFs, we looked to see if 5′ tRFs affect mRNA recognition by eIF4F and/or poly(A) binding protein (PABP). In the early stages of translation, the eIF4F complex binds the mRNA cap structure and PABP binds the poly(A) tail. Interactions between eIF4F and PABP cause mRNA circularization, which is required for efficient translation initiation.Citation35 In our in vitro system, uncapped or non-adenylated mRNA can be translated, although at low efficiency. We investigated whether Gln19 caused a translation defect from mRNA that is either capped but non-adenylated, or which contains an “ACap” (that cannot interact with eIF4F) and is non-adenylated. As shown in , we found that translation of these mRNAs was inhibited to the same degree as for capped, polyadenylated mRNA, indicating that translation inhibition does not occur by interfering with the mRNA-circularization steps of initiation.
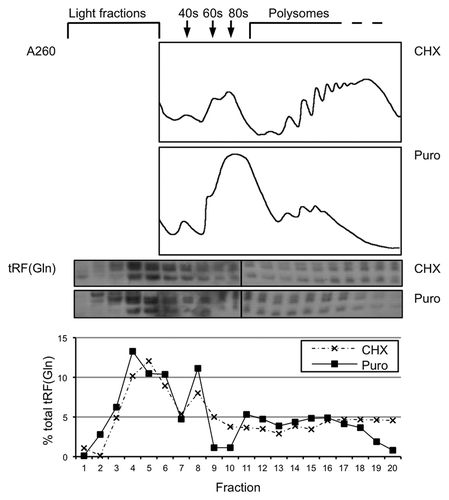
We next investigated whether tRFs are associated with particular translation intermediates by separating sub-ribosomal complexes from polysomes on a sucrose gradient. In cells treated with cycloheximide to freeze polysomes, most 5′ tRFs were recovered from the low density fractions, but a significant proportion of cellular tRF(Gln) comigrated with polysomes (, upper northern blot). When translationally active polysomes were disrupted by addition of the chain terminator puromycin to cells prior to lysis, the distribution of tRF(Gln) shifted toward lighter fractions concomitantly to the A260 trace (, lower northern blot; also compare quantification of fractions 17–20). In the highest-occupancy polysomes, represented by fractions 17–20, the amount of tRF(Gln) recovered was 18.5% in the cyclohexamide-treated cells but 10.5% in the puromycin-treated cells where these polysomes had been specifically disrupted. This indicates that 5′ tRFs are associated with bona-fide polysomes and that these polysomes are not permanently stalled translation complexes. We also investigated whether the biochemical distribution of a target mRNA changed after cotransfection with tRF sequences. To do this, we co-transfected cells with psiCheck-2 [in order to express Renilla luciferase, a known target of tRF(Gln)] and Gln19. We observed similar amounts of Renilla mRNA present in polysomes in samples transfected with Gln19, Gln19(mGG) or no RNA, indicating that formation of elongating ribosomes does not appear to be affected by 5′ tRF sequences (Fig. S5).
Figure 8. A proportion of cellular tRFs associate with polysomes. Cell lysate from HeLa cells was separated on a 10–50% (w/v) sucrose gradient to resolve polysomes. Cells were either treated with 50 µg/mL cycloheximide (7 min) or the chain terminator puromycin (200 µg/mL, 15 min) before lysis. The absorbance profile at 260 nm is shown. tRF(Gln) was detected using northern blotting, and quantifications of the level in each fraction are shown below the blots.
Discussion
In addition to their well-characterized function as amino acid carriers, it has recently become apparent that tRNAs are used as precursors for a number of different classes of small RNAs.Citation22 They can be cleaved by various endonucleases into 3′ U, 3′ CCA or 5′ tRFs, and they can be cleaved by angiogenin into half-tRNA fragments under stress conditions. tRNA fragments have been found to have a variety of different functions: some 5′ tRNA halves specifically inhibit translation by a mechanism involving sequestration of eIF4G;Citation27 some 3′ tRFs have been shown to have functions in cell proliferation;Citation14,Citation23 and 3′ U and 3′ CCA tRFs have been shown to associate with the small RNA effector machinery.Citation17
In our previous work, we showed that a 5′ tRF, tRF(Gln), is only weakly associated with Argonaute proteins,Citation12 suggesting that it may function outside of the canonical small RNA pathways. Here, we show that it does not have significant siRNA/miRNA-like functions in a suitable reporter assay system. However, synthetic RNAs containing 5′ tRF sequences are able to repress the expression of two different reporter genes, in a process that does not require the existence of target sites in the mRNA.
These data show notable parallels to a recent study that finds that tRNA fragments have a direct inhibitory effect on protein translation in the archeon Haloferax volcanii.Citation36 In this study, Gebetsberger et al. show that 26 nt 5′ tRFs are able to inhibit translation. Specifically, they find that tRF(Val) is able to inhibit peptide bond formation, and that another, less abundant tRF [tRF(Ile)] is not. The fact that 5′ tRFs are able to inhibit translation across domains of life suggest that this may be an ancient way by which organisms control protein translation. Out of the four RNAs that we tested, three of them (derived from different tRNAs) were able to inhibit translation, and in these tRFs the activity required a conserved “GG” dinucleotide. Indeed, a non-tRNA-derived sequence could inhibit translation in the same manner as 5′ tRF sequences if a 3′ “GG” dinucleotide was present. However, we note that one tested RNA molecule (Glu19) did not cause any translational repression, indicating that perhaps other sequence features can affect the ability of these RNA molecules to modulate translation. It is noteworthy that Gebetsberger et al. also find that only a subset of 26-nt 5′ tRFs are able to inhibit translation.Citation36
5′ tRNA halves also inhibit translation, but by a different mechanism.Citation27 5′ tRNA halves are derived from the same part of tRNA molecules as 5′ tRFs, indeed they contain the 5′ tRF sequence. However, translation repression by 5′ tRNA halves requires a run of at least four guanosine residues at the 5′ end of the molecule, which is present in tRNAAla and tRNACys only. In contrast, 5′ tRFs require only two guanosine residues at the 3′ end of the molecule, and these residues are absolutely conserved between tRNAs.Citation27 Interestingly, although 5′ tRNA halves contain the 5′ tRF sequence, the mutation of the invariant “GG” dinucleotide at positions 17-18/18-19 does not affect their ability to repress translation,Citation27 which contrasts to the 5′ tRFs we have investigated. Additionally, 5′ tRFs and 5′ tRNA halves are probably produced by different pathways, as tRNA halves are produced in response to different stimuli to 5′ tRFs. tRNA halves are known to be produced when cells experience oxidative stress,Citation19 but under conditions of oxidative stress we observe no change in 5′ tRF levels (data not shown). Instead, 5′ tRF levels appear to be elevated in cells exhibiting a high proliferation rate.
We have shown that the inhibition of protein translation by 5′ tRFs is independent of mRNA circularization. In addition, the mRNA abundance and polyadenylation status is not affected by 5′ tRFs, and nor is the abundance or charge state of a cognate tRNA. The exact mechanism of inhibition remains to be elucidated in further work. Our observation that a fraction of cellular 5′ tRFs associate with actively elongating polysomes, taken together with the fact that mRNA association with polyribosomes does not change when 5′ tRFs are transfected into cells, may indicate that they interfere with the process of elongation. However, we note that the majority of 5′ tRFs are present in complexes smaller than the 40S small ribosomal subunit and, hence, they may alternatively exert their effects at other points in the translation cycle. As well as the canonical translation control points, they could also interfere with processes such as the specificity of tRNA charging; since our measurements of translation are based on functional enzyme assays, we would also detect defects in translation fidelity caused by mischarging of tRNAs, as has recently been reported by Netzer et al.Citation37
In the light of these data, what is the function of 5′ tRFs in vivo? While we note that our experiments to date have been focused on the effect of 5′ tRFs on translation of a limited number of reporter genes, and we do not claim that 5′ tRFs would affect all genes equally, it nevertheless seems apparent that they repress the translation of at least a subset of mRNAs. Cell growth and division requires a high translation rate, and the growth of tumors requires mutations that increase bulk cellular translation rate.Citation38 Some 3′ U and 5′ tRFs have been shown to be elevated in actively proliferating cells.Citation14,Citation17 Maybe 5′ tRFs represent a mechanism by which cells can put the brakes on hyperactive translation? A 3′ CCA tRF that acts as a miRNA suppresses proliferation by targeting the RPA1 gene.Citation23 The interesting observation that a 3′ U tRF, tRF-1001, is necessary for cell growthCitation14 leads to the hypothesis that cells finely balance their translational output and/or proliferation by modulation of the production or stability of different types of tRNA fragment. The investigation of these and similar questions in suitable cellular contexts will lead to a greater understanding of these new types of RNA.
Materials and Methods
Oligonucleotide sequences
RNA oligonucleotides were as follows: Gln19, GGUUCCAUGGUGUAAUGGU; Gln19(Scr), GUGGUAGAGGUUGCUAUCU;; Gln19(mMid), GGUUCCAACCUGUAAUGGU; Gln19(mGG), GGUUCCAUGGUGUAAUCCU; Glnmid, GAGCACUUUGGACUCUGAA; Val19, GUUUCCGUAGUGUAGUGGU; Val19(mGG), GUUUCCGUAGUGUAGUCCU; Lys19, GCCCGGAUAGCUCAGUUGG; Lys19(mGG), GCCCGGAUAGCUCAGUUCC; Glu19, UCCCUGGUGGUCUAGUGGU; Artificial19, CAUCCAUAAGUCGCAUGGU; Artificial19(mGG), CAUCCAUAAGUCGCAUCCU; Artificial19(unstruct), AAUAAAUAAGUCGCAUGGU; Artificial19(loopswap), CAUCCAUGUACCUAAUGGU; tRF-1001, GAAGCGGGUGCUCUUAUUU; siGln, GGUUCCAUGGUGUAAUGGUdTdT (sense), ACCAUUACACCAUGGAACAdTdT (passenger). DNA oligonucleotides were as follows: let-7a_target, GTTGCGGCCGCTGAGGTAGTAGGTTGTATAGTTTCGACTGAGGTAGTAGGTTGTATAGTTTCGACTGAGGTAGTAGGTTGTATAGTTCTCGAGTTG;; let-7a_mut_target, GTTGCGGCCGCTGTCCTAGTAGCTTGTATAGTTTCGACTGTCCTAGTAGCTTGTATAGTTTCGACTGTCCTAGTAGCTTGTATAGTTCTCGAGTTG; gln_target, GTTGCGGCCGCGGTTCCATGGTGTAATGGTTCGACGGTTCCATGGTGTAATGGTTCGACGGTTCCATGGTGTAATGGTCTCGAGTTG;; gln_mut_target, GTTGCGGCCGCGGAAGCATGGTCTAATGGTTCGACGGAAGCATGGTCTAATGGTTCGACGGAAGCATGGTCTAATGGTCTCGAGTTG; let-7a_ds, CAACTCGAGAACTATACAA; gln_ds, CAACTCGAGACCATTACAC; gln_mut_ds, CAACTCGAGACCATTAGAC; glnmm_XPE, ATCGAACTCGAGCAGGGTTTAAACACCATTAGTGGATGGAACCCAGGACCATTAGTGGATGGAACCCAGGACCATTAGTGGATGGAACCGATATCCTCGAA; glnmm_mut_XPE, ATCGAACTCGAGCAGGGTTTAAACACCATTAGTGGATGCTTCCCAGGACCATTAGTGGATGCTTCCCAGGACCATTAGTGGATGCTTCCGATATCCTCGAA; m5_mut_fwd, CCTCCAGCTACCTCTGGCGCCACGTCGTGCCTC; m5_mut_rev, TTCTCCTTGAATGGTTCGAGGTAGGCAGCGAACTCC; m51_mut, TTCGCTGCCTACCTCGAGCCATTCAAGGAGAA. The following qPCR primers were used: Renilla, ACAAGTACCTCACCGCTTGG (fwd).
RNA secondary structure prediction
RNA structures were predicted by the Mfold software.Citation39
Plasmid construction
To generate psiCheck-3xGln(perfect and perfect_control) and psiCheck-3xlet-7(perfect and perfect_control), an oligonucleotide (let-7a_target, let-7a_mut_target, gln_target, gln_mut_target) containing the target sites was made double stranded using an appropriate primer (let-7a_ds, gln_ds, gln_mut_ds) and Klenow's reagent. These inserts were digested with NotI and XhoI and ligated into psiCheck-2 (Promega) that had been digested with the same restriction enzymes and 3′ dephosphorylated.
To generate psiCheck-6xGln(mismatch and mismatch_control), an oligonucleotide containing 3 target sites (glnmm_XPE and glnmm_mut_XPE) were made double stranded by incubation with their reverse complement at 95°C followed by cooling to room temperature. These inserts were digested with XhoI and EcoRV and ligated into psiCheck-2 that had been digested with XhoI and PmeI and 3′ dephosphorylated. This protocol was repeated to add another three target sites. During cloning, the PmeI site was modified such that psiCheck-6xGln(mismatch) had sequence GATCGCTCGACAGGGTAAAC and psiCheck-6xGln(mismatch_control) had sequence GATCGCTCGAGCAGGGTTAAAC.
To create psiCheck-2(m5), psiCheck-2 was mutagenized in a standard mutagenic PCR reaction in a final volume of 50 µL containing 125 ng primers m5_mut_fwd and m5_mut_rev, 200 nM dNTPs, 25 ng psiCheck-2 and 2.5 U PfuTurbo (Stratagene) (cycle settings: initial denaturing stage 95°C 30s, then 95°C 30s, 55°C 1 min, 68°C 6.5 min, for 18 cycles). The product was desalted using QIAquick columns (Qiagen), and eluted in a volume of 26 µL. The template DNA was digested by incubation with 1U FastDigest DpnI (Fermentas) at 37°C for 20 min. Two µL of the reaction mixture was used for transformation of E. coli (XL1 Blue).
psiCheck-2(m51) was created in a similar way from psiCheck-2(m5) by mutagenesis with primer m51_mut and its reverse complement.
All constructs generated were verified by appropriate sequencing (DNA sequencing service, University of Dundee).
Cell culture and transfection
HeLa cells were cultured in DMEM containing 10% (v/v) fetal calf serum, 25 U/mL penicillin and 25 µg/mL streptomycin (Invitrogen). Transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
In vivo luciferase reporter assays
HeLa cells in a 24-well plate format were transfected with 0.13 µg reporter plasmid. Where siRNA or 2′ O-methyl oligonucleotides were co-transfected, 40 pmol was used, and where unmodified RNA was cotransfected, 100 pmol was used. Sixteen hours after transfection, cells were lysed using passive lysis buffer (Promega) and luciferase activity measured using the Dual-Luciferase Reporter Assay System (Promega) in accordance with the manufacturer's instructions.
Calculation of relative Renilla expression
The relative Renilla expression when cells were transfected with a scrambled antisense RNA compared with a tRF(Gln) antisense RNA () was calculated as follows:
where and
are the raw expression levels of Renilla and firefly luciferases, respectively.
In vitro translation
Renilla mRNA was generated by T7 in vitro transcription of psiCheck-2 (Promega) that had been linearized with NotI. The RNA transcript was polyadenylated using E. coli poly(A) polymerase (NEB). In vitro translation reactions were performed as described in reference Citation30 with the following concentrations of factors in 10 µL reactions: 16 mM HEPES-KOH pH 7.6, 75 mM KOAc, 2.5 mM Mg(OAc)2, 800 µM ATP, 100 µM GTP, 125 µM spermidine (Sigma Aldrich), 100 µM amino acids, 25 mM creatine phosphate (Sigma Aldrich), 80 ng/µL creatine kinase (Sigma Aldrich), 100 ng/µL tRNA (Sigma Aldrich), 1 µL/10 µL RNasin (Promega), 10 ng/µL mRNA and 2 µL HeLa cytoplasmic extract per reaction. Reactions were performed at 37°C for 1 h. Renilla activity was subsequently assayed using the Renilla Luciferase Assay System (Promega).
qPCR
RNA was always treated with DNA-free (Applied Biosystems) before analysis by qPCR. Reverse transcription of 200 ng RNA was performed with Superscript III (Invitrogen) in accordance with the manufacturer's instructions, using either random hexamers or dT20 as noted in figure legends. Six perecent of this reaction was then used for qPCR. qPCR was performed using SYBR Green master mix (Qiagen) in accordance with the manufacturer's instructions in a 25 µL reaction volume, and monitored on an Applied Biosystems 7500 Real-Time PCR System. The dissociation curve was monitored for primer specificity and controls with non-reverse-transcribed RNA were always performed.
tRNA aminoacylation assay
To assay tRNA aminoacylation state, RNA was extracted using Trizol reagent (Invitrogen) but the sample was kept on ice at all stages to avoid deacylation. The final RNA was resuspended in 10 mM NaOAc pH 5.0. To obtain a deacylated marker, a fraction of this RNA was resuspended in 200 mM Tris pH 9.5 and incubated at 37°C for 60 min.
To separate aminoacylated and non-aminoacylated RNA, a 6.5% gel (19:1 acrylamide:bis) was cast in 100 mM NaOAc pH 5.0 and run in the same buffer at 4°C. This gel was blotted onto a positively charged nitrocellulose membrane (GE Healthcare), crosslinked by exposure to UV light (254 nm, 120,000 µJ/cm2) and probed with complementary RNA oligonucleotides.
Sucrose gradient analysis
Cells were lysed on ice in a buffer containing 10 mM Tris-Cl (pH 8.0), 0.5% (v/v) NP-40 (Calbiochem), 130 mM KCl, 10 mM MgCl2, 10 µg/mL cycloheximide (Sigma Aldrich), 200 µg/mL heparin (Sigma Aldrich), 2.5 mM DTT, 0.2 U/µL RNasin (Promega) and complete protease inhibitor (Roche). After 20 min lysis, soluble material was isolated by centrifugation and loaded onto a sucrose gradient of 10–50% (v/v) sucrose in 200 mM Tris-Cl pH 8.0, 50 mM KCl, 25 mM MgCl2, 1.5 mM DTT, contained in a 13.2 mL polyallomer tube (Beckman, catalog #331372). This gradient was centrifuged in an SW41-Ti rotor (36,000 rpm, 4°C, 3 h) and fractions collected and A260 monitored.
Additional material
Download Zip (722 KB)Acknowledgments
This work was supported by the European Framework 6 SIROCCO, the Wellcome Trust Centre for Gene Regulation and Expression and the ARC DP 130103027. G.H is an ARC Future Fellow.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Farazi TA, Juranek SA, Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development 2008; 135:1201 - 14; http://dx.doi.org/10.1242/dev.005629; PMID: 18287206
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001; 409:363 - 6; http://dx.doi.org/10.1038/35053110; PMID: 11201747
- Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 2001; 293:834 - 8; http://dx.doi.org/10.1126/science.1062961; PMID: 11452083
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 2007; 128:1089 - 103; http://dx.doi.org/10.1016/j.cell.2007.01.043; PMID: 17346786
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 2004; 15:185 - 97; http://dx.doi.org/10.1016/j.molcel.2004.07.007; PMID: 15260970
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science 2004; 305:1437 - 41; http://dx.doi.org/10.1126/science.1102513; PMID: 15284456
- Pillai RS, Artus CG, Filipowicz W. Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA 2004; 10:1518 - 25; http://dx.doi.org/10.1261/rna.7131604; PMID: 15337849
- Ender C, Krek A, Friedländer MR, Beitzinger M, Weinmann L, Chen W, et al. A human snoRNA with microRNA-like functions. Mol Cell 2008; 32:519 - 28; http://dx.doi.org/10.1016/j.molcel.2008.10.017; PMID: 19026782
- Bühler M, Spies N, Bartel DP, Moazed D. TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat Struct Mol Biol 2008; 15:1015 - 23; http://dx.doi.org/10.1038/nsmb.1481; PMID: 18776903
- Kawaji H, Nakamura M, Takahashi Y, Sandelin A, Katayama S, Fukuda S, et al. Hidden layers of human small RNAs. BMC Genomics 2008; 9:157; http://dx.doi.org/10.1186/1471-2164-9-157; PMID: 18402656
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev 2008; 22:2773 - 85; http://dx.doi.org/10.1101/gad.1705308; PMID: 18923076
- Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, et al. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 2009; 15:2147 - 60; http://dx.doi.org/10.1261/rna.1738409; PMID: 19850906
- Yeung ML, Bennasser Y, Watashi K, Le S-Y, Houzet L, Jeang K-T. Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res 2009; 37:6575 - 86; http://dx.doi.org/10.1093/nar/gkp707; PMID: 19729508
- Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev 2009; 23:2639 - 49; http://dx.doi.org/10.1101/gad.1837609; PMID: 19933153
- Hsieh L-C, Lin S-I, Shih AC-C, Chen JW, Lin WY, Tseng CY, et al. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol 2009; 151:2120 - 32; http://dx.doi.org/10.1104/pp.109.147280; PMID: 19854858
- Liao J-Y, Ma L-M, Guo Y-H, Zhang YC, Zhou H, Shao P, et al. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3′ trailers. PLoS One 2010; 5:e10563; http://dx.doi.org/10.1371/journal.pone.0010563; PMID: 20498841
- Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 2010; 16:673 - 95; http://dx.doi.org/10.1261/rna.2000810; PMID: 20181738
- Couvillion MT, Sachidanandam R, Collins K. A growth-essential Tetrahymena Piwi protein carries tRNA fragment cargo. Genes Dev 2010; 24:2742 - 7; http://dx.doi.org/10.1101/gad.1996210; PMID: 21106669
- Yamasaki S, Ivanov P, Hu G-F, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol 2009; 185:35 - 42; http://dx.doi.org/10.1083/jcb.200811106; PMID: 19332886
- Li Z, Ender C, Meister G, Moore PS, Chang Y, John B. Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs. Nucleic Acids Res 2012; 40:6787 - 99; http://dx.doi.org/10.1093/nar/gks307; PMID: 22492706
- Pederson T. Regulatory RNAs derived from transfer RNA?. RNA 2010; 16:1865 - 9; http://dx.doi.org/10.1261/rna.2266510; PMID: 20719919
- Sobala A, Hutvagner G. Transfer RNA-derived fragments: origins, processing, and functions. Wiley Interdiscip Rev RNA 2011; 2:853 - 62; http://dx.doi.org/10.1002/wrna.96; PMID: 21976287
- Maute RL, Schneider C, Sumazin P, Holmes A, Califano A, Basso K, et al. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci USA 2013; 110:1404 - 9; http://dx.doi.org/10.1073/pnas.1206761110; PMID: 23297232
- Lee SR, Collins K. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J Biol Chem 2005; 280:42744 - 9; http://dx.doi.org/10.1074/jbc.M510356200; PMID: 16272149
- Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 2008; 14:2095 - 103; http://dx.doi.org/10.1261/rna.1232808; PMID: 18719243
- Emara MM, Ivanov P, Hickman T, et al. Angiogenin-induced tiRNAs promote stress-induced stress granule assembly. J Biol Chem 2010; 285:10959 - 68; http://dx.doi.org/10.1074/jbc.M109.077560; PMID: 20129916
- Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell 2011; 43:613 - 23; http://dx.doi.org/10.1016/j.molcel.2011.06.022; PMID: 21855800
- Schwarz DS, Hutvágner G, Haley B, Zamore PD. Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol Cell 2002; 10:537 - 48; http://dx.doi.org/10.1016/S1097-2765(02)00651-2; PMID: 12408822
- Hutvágner G, Simard MJ, Mello CC, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol 2004; 2:E98; http://dx.doi.org/10.1371/journal.pbio.0020098; PMID: 15024405
- Bergamini G, Preiss T, Hentze MW. Picornavirus IRESes and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. RNA 2000; 6:1781 - 90; http://dx.doi.org/10.1017/S1355838200001679; PMID: 11142378
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA 2004; 10:1507 - 17; http://dx.doi.org/10.1261/rna.5248604; PMID: 15383676
- Marck C, Grosjean H. tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA 2002; 8:1189 - 232; http://dx.doi.org/10.1017/S1355838202022021; PMID: 12403461
- Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 2011; 12:99 - 110; http://dx.doi.org/10.1038/nrg2936; PMID: 21245828
- Varshney U, Lee CP, RajBhandary UL. Direct analysis of aminoacylation levels of tRNAs in vivo. Application to studying recognition of Escherichia coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J Biol Chem 1991; 266:24712 - 8; PMID: 1761566
- Tarun SZ Jr., Sachs AB. Binding of eukaryotic translation initiation factor 4E (eIF4E) to eIF4G represses translation of uncapped mRNA. Mol Cell Biol 1997; 17:6876 - 86; PMID: 9372919
- Gebetsberger J, Zywicki M, Künzi A, Polacek N. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea 2012; 2012:260909; http://dx.doi.org/10.1155/2012/260909; PMID: 23326205
- Netzer N, Goodenbour JM, David A, Dittmar KA, Jones RB, Schneider JR, et al. Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature 2009; 462:522 - 6; http://dx.doi.org/10.1038/nature08576; PMID: 19940929
- Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nat Rev Cancer 2010; 10:254 - 66; http://dx.doi.org/10.1038/nrc2824; PMID: 20332778
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 2003; 31:3406 - 15; http://dx.doi.org/10.1093/nar/gkg595; PMID: 12824337