Abstract
Post-synthetic modifications of nucleic acids have long been known to affect their functional and structural properties. For instance, numerous different chemical modifications modulate the structural organization, stability or translation efficiency of tRNAs and rRNAs. In contrast, little is known about modifications of poly(A)RNAs. Here, we demonstrate for the first time that the two well-studied regulatory long non-coding RNAs HOTAIR and XIST are targets of site-specific cytosine methylation. In both XIST and HOTAIR, we found methylated cytosines located within or near functionally important regions that are known to mediate interaction with chromatin-associated protein complexes. We show that cytosine methylation in the XIST A structure strongly affects binding to the chromatin-modifying complex PRC2 in vitro. These results suggest that cytosine methylation may serve as a general strategy to regulate the function of long non-coding RNAs.
Introduction
The presence of chemical modifications of nucleic acids has been firmly established over 60 y ago with the discovery of 5-methylcytidine.Citation1,Citation2 Over the years, it became evident that various chemical groups can be added to or removed from sugar and base moieties of DNA and RNA.Citation3 In DNA, the best-studied modification is the methylation of the carbon C5 of cytosine (m5C), which plays an important role in gene regulation and the epigenetic memory of a cell.Citation4
In RNA, post-transcriptional modifications, in particular methylation, are much more complex because they can affect all bases as well as the ribose moiety, and they can occur in a highly dynamic fashion.Citation3,Citation5,Citation6 The two most abundant cellular RNA species, rRNA and tRNA, have been studied in detail and the exact positions of various methylations have been determined.Citation5,Citation7 Nevertheless, the functional or biological consequences of many post-transcriptional modifications in RNA, including base methylation, remain largely elusive.
While the types and positions of methylation in rRNA and tRNA are well characterized,Citation3 much less is known about methylation in poly(A)RNA. Work from the 1970s demonstrated that the majority of internal methylations are on nitrogen 6 of adenosines (m6A),Citation8,Citation9 whereas m5C was found to constitute a small fraction of methylated mRNA in hamster BHK-21 and HeLa cellsCitation8,Citation9 but was not detected in Novikoff hepatomaCitation10 or mouse myeloma cells.Citation11 Only recently a transcriptome-wide picture of m6A was gained by employing antibody enrichment combined with deep sequencing methods.Citation12,Citation13 Likewise, during the preparation of this manuscript, a global analysis of m5C levels in HeLa cells was published, showing the widespread distribution of this modification in mRNA.Citation14 One reason for the huge gap between the initial detection of these mRNA modifications and their further study was the lack of suitable methods. While bisulfite (BS) sequencing is used as a robust method to assess m5C in DNA,Citation15 application of this technique to RNA was hampered by the greater instability of phosphodiester bonds at high temperature and high pH, which leads to pronounced RNA degradation. Recently, it has been possible to adapt the BS treatment protocol for the analysis of m5C in the abundant rRNAs and tRNAs.Citation16
In the present study, we sought to explore the presence of site-specific methylation of cytosines in less abundant cellular RNAs. Similar to the method published by Squires et al.,Citation14 we developed a protocol based on bisulfite treatment and PCR amplification of poly(A)-enriched RNA to investigate the possibility of cytosine methylation in the long non-coding (lnc) RNAs HOTAIR and XIST.
Results
Discovery of m5C in HOTAIR RNA
The function of m5C in tRNA and rRNA has been mainly linked to translation controlCitation17 and RNA stability.Citation18-Citation20 In addition, there is evidence for a structural function of m5C in tRNA.Citation5,Citation21 Since at least some lncRNAs are known to form extended secondary and tertiary structures,Citation22,Citation23 we reasoned that they might represent promising candidates in the search for cytosine methylation among poly(A)RNAs. Several species, such as the mammalian XIST or HOTAIR RNAs, have been suggested to function at the chromatin level by interacting with chromatin modifying protein complexes.Citation23-Citation25 This ability may involve structural rather than sequence features of the respective RNAs. We hypothesized that if cytosine methylation affects higher-order structure and possibly protein-interaction capabilities of an RNA, then m5C sites would most likely be found within functionally important domains.
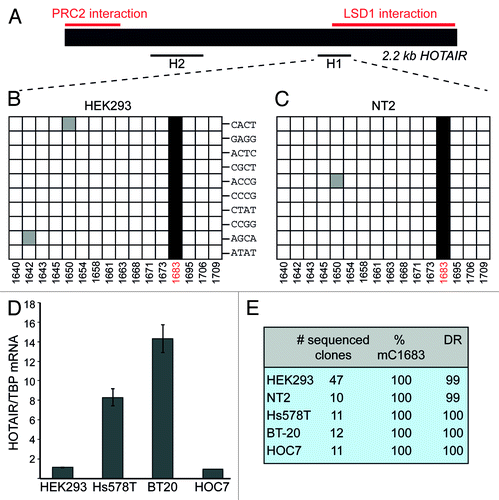
We selected the HOTAIR lncRNA, which has previously been shown to interact directly with chromatin-modifying complexes,Citation24 to investigate the presence of m5C by bisulfite sequencing. To increase the chances for detection of HOTAIR RNA after BS treatment, we adapted the method of Schaefer et al.Citation16 by starting with poly(A)-enriched nuclear RNA (Fig. S1). A region of ~300 nt at the 5′-end of HOTAIR has been described to associate with the polycomb-repressive complex 2 (PRC2), whereas an approximately 700 nt sequence at the 3′-end interacts with the histone demethylase complex LSD1/CoREST/REST.Citation23,Citation24 Restricted by the difficulties of primer design for deaminated sequences, we were nevertheless able to analyze a short region in the vicinity of the LSD1-binding site (H1) as well as another region in the middle of the sequence (H2) (). The analysis of the H2 region revealed no methylated Cs (Fig. S2). In contrast, in fragment H1 we discovered a methylated C at position 1683 that was present in all sequenced clones derived from HEK293 cells (). To confirm that the observed complete penetrance of C1683 methylation was not due to biased amplification of non-deaminated cDNA molecules, we used bar-coded primers for a single round of hexamer-primed cDNA labeling, before PCR amplification of the H1 product. shows that each analyzed clone contained a different barcode label, which argues against a significant PCR bias. Next we asked whether methylation of C1683 was cell type-specific. We analyzed RNA isolated from the human NT2 cell line. Once again, all examined clones displayed C methylation at position 1683 ().
Figure 1. HOTAIR lncRNA shows m5C in various cell types. (A) Schematic representation of HOTAIR RNA. Previously mapped PRC2 and LSD1-interacting regions are marked in red. Black lines designate the regions that were analyzed by BS sequencing. (B and C) Sequencing results of 10 clones derived from BS treated HOTAIR RNA from HEK293 (B) and NT2 (C) cells. Each horizontal row in the diagrams corresponds to one sequenced clone, each square to a specific C (numbers at the bottom indicate the position of the C within the HOTAIR sequence). Non-deaminated (i.e. methylated) Cs are shown as black squares, white squares are deaminated Cs and gray squares depict non-deaminated Cs that were considered BS treatment artifacts. Barcode labels of individual clones are indicated in (B). (D) Expression analysis of HOTAIR in the breast cancer cell lines Hs578T, BT20 and in HOC7 ovarian cancer cells. RT-qPCR results are expressed relative to values obtained from HEK293 cells. (E) Summary of BS sequencing results for HOTAIR in different cell lines; DR, deamination rate in percent.
HOTAIR methylation is not altered in cancer cells
HOTAIR has recently gained increased attention for its putative role in promoting metastatic activity of several cancer types.Citation23 Since it was shown that increased HOTAIR expression in cancer cells correlated with poor prognosis for the patients, we investigated whether C1683 methylation is affected by the elevated amount of HOTAIR transcript in the Hs578T and BT-20 breast cancer and the HOC7 ovarian cancer cell lines (). BS sequencing analysis of HOTAIR showed that C1683 was invariably methylated in the three cancer cell lines () in spite of considerably different HOTAIR expression levels ().
These results demonstrate that there is site-specific cytosine methylation in the lncRNA HOTAIR. Methylation of C1683 is widespread in different cell types and it is not limited by the abundance of HOTAIR RNA levels. Furthermore, the degree of methylation of C1683 appears to be remarkably high, suggesting that it might be important for the function of HOTAIR. In this respect, it is interesting to note that C1683 is located in the vicinity of the region that has previously been shown to interact with the LSD1 complex.Citation23,Citation24 It is therefore tempting to speculate that methylation of this cytosine may affect the ability of HOTAIR to interact with LSD1.
XIST RNA is methylated at specific cytosines within the A-region
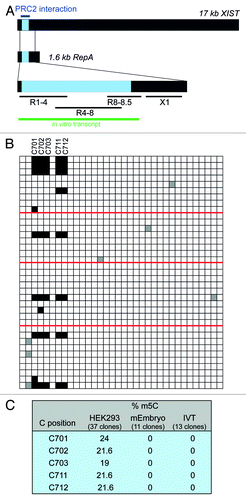
We then examined one of the best-studied human lncRNAs, the XIST RNA, for m5C methylation. XIST is required for the permanent inactivation of one of the two X chromosomes in female mammals by coating the chromosome in cis.Citation26 The mechanism of silencing involves association with the PRC2 complex via a region at the 5′-end of XIST, termed the repeat A-region.Citation26 This functionally important domain is composed of 8.5 repeats (R1-R8.5) of a 26 nt sequence. To determine if the A-region could be a target of C methylation, we designed primers to scan the entire A-region and an adjacent downstream segment for possible m5C sites by BS sequencing of RNA from the female HEK293 cell line (). Sequence analysis of at least 10 clones each of fragments R1-4, R4-8 and fragment X1 did not reveal enhanced protection of particular cytosine residues from bisulfite induced deamination (Fig. S3B‒D). In contrast, analysis of region R8-8.5 showed distinct retention of a cluster of cytosines within R8 in several clones. In a total of 37 sequences derived from multiple experiments, we observed methylation at cytosines 701, 702, 703, 711 and 712 in 19‒24% of the clones. Simultaneous methylation of all five cytosines was observed in 19% of the sequences (). Although we attempted the use of barcoded primers to rule out a PCR bias toward non-deaminated sequences, we were unable to obtain PCR products with these primers. However, in light of the HOTAIR results (), the likelihood of a bias introduced by PCR seems small.
Figure 2. Cytosine methylation within A-region repeat 8 of XIST. (A) Schematic representation of XIST RNA and the overlapping RepA transcript. The A region is shown in blue. Blue line, PRC2 interaction domain; black lines, regions analyzed by BS sequencing; green line, size and position of the A-region in vitro transcript. (B) BS sequencing results for region R8-8.5. Red lines demarcate results obtained from different experiments with fresh batches of cells. (C) Summary of BS sequencing results of R8-8.5 in different cell types as well as in in vitro transcribed (IVT) XIST templates.
We next examined if XIST methylation is conserved also in the A-region of mouse XIST. To this end, we analyzed RNA from a female mouse embryo at E17. Although the sequences of mouse repeat 7 (which corresponds to repeat 8 in human XIST) are virtually identical, no methylation was detected at the corresponding cytosines C668, 669, 670 and 678 (Fig. S3E).
Since the degree of cytosine methylation at the detected positions was considerably lower in XIST than in HOTAIR, we sought to rule out the possibility that enhanced protection of these cytosines was due to potential rigid secondary and tertiary structure formation in this region, which would inhibit bisulfite-induced cytosine deamination. To this end, the entire A-region was transcribed in vitro, and the resulting RNA was heat-denatured and then slowly cooled to promote RNA folding. To mimic the complexity of a natural sample, the XIST transcript was mixed with an excess of total E. coli RNA prior to BS treatment. Sequence analysis of individual clones derived from this RNA revealed complete conversion of all the cytosines in question to uracil (). Therefore, given (1) the high overall deamination rate of > 95% in our experiments (Fig. S3), (2) the detection of C methylation in R8 but not in any of the other repeats containing identical sequences, (3) the observed species and/or cell type-specificity of the observed methylation and (4) the fact that in vitro transcribed and, thus, non-methylated RNA showed complete C deamination, we conclude that C701-712 are genuine methylation targets in XIST.
Potential relationship between cytosine methylation and functional properties of XIST RNA
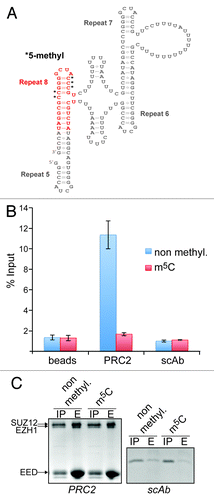
A key question that arises with the discovery of m5C in lncRNAs, such as HOTAIR and XIST, is what consequences this methylation might have for the function of the RNAs. We have begun to investigate this issue by assessing the position of the methylated cytosine(s) in the context of their structural surroundings. In this analysis, we focused on XIST because the structural organization of its A-region has been studied in greater detail. Three-dimensional structure modeling revealed a distinctive propensity of R8 to enter in helical stacking interactions (see supplementary results and Figs. S4 and S5 for a more detailed description). Furthermore, in this conformation, all the discovered methylated cytosines are positioned in close proximity on the external section of the helix (Fig. S5). Several reports have shown that the XIST A-region can interact with PRC2, although there remains some ambiguity as to the exact nature of the interaction.Citation25,Citation27-Citation29 Nevertheless, it is possible that methylation of specific Cs in the hairpin stem may modulate the interaction surface between XIST and the PRC2 complex
Cytosine methylation affects the binding properties of R8 to the PRC2 complex
To test this hypothesis, we performed in vitro RNA-binding assays. We synthesized an RNA oligonucleotide spanning the R8 tetraloop sequence and part of the inter-repeat helix () and bearing methyl groups on C701, 702, 703, 711 and 712. We then analyzed the ability of recombinant Flag-tagged PRC2 to interact with radiolabeled methylated and unmethylated R8 RNA by measuring retention of radioactivity on anti-Flag agarose beads. These experiments revealed that PRC2 was able to bind to the unmodified R8 RNA (). In striking contrast, no significant retention of radioactivity was detected when the methylated RNA was used (). The absence of retention of the radiolabeled methylated RNA was not due to an interference with the binding of the PRC2 complex to the beads (). These data clearly indicate that cytosine methylation in R8 of XIST has the potential to modulate RNA-protein interactions suggesting that posttranscriptional modification of nucleotides may be a mechanism to regulate the function of lncRNAs.
Figure 3. Cytidine methylation in XIST repeat 8 prevents binding of the PRC2 complex. (A) Two-dimensional structure model of R5-R8 showing the position and sequence of the RNA used in the RNA-binding assay (marked in red). Black asterisks indicate the methylated cytidines. (B) Bar graph showing the quantification by scintillation counting of 32P-5′ end-labeled XIST R8 RNA bound to affinity-purified PRC2. Flag-tagged PRC2 or a Flag-tagged control protein (scAb) was incubated with either the unmodified RNA (blue bars) or RNA methylated on the 5 cytidines indicated in (A) (red bars) and subsequently purified by Flag-M2-agarose. In addition, the RNAs were incubated with anti-Flag-beads in the absence of protein (beads). Values represent mean +/− SD of three independent experiments. (C) Coomassie blue stained SDS-polyacrylamide gels showing input material (IP) and eluates (E) of Flag-M2-beads from reactions described in (B) for PRC2 (left panel) and scAb (right panel). Twenty-five percent and 100%, respectively, of input of PRC2 and scAb were loaded in the IP lanes, 30% of eluates were loaded in the E lanes. The positions of the PRC2 subunits EZH1 (86 kDa), SUZ12 (87 kDa) and EED (51 kDa) are indicated. The MW of scAb is 18 kDa.
Discussion
In this study we show that the two lncRNAs, HOTAIR and XIST, are targets for site-specific cytosine methylation in vivo and that, at least in the case of XIST, this modification has the ability to affect protein binding. It is important to note that the BS sequencing technique may not be able to distinguish between several different cytosine modifications, such as N4,2’-O-dimethylcytidine (m4mC)Citation16 or 5-hydroxymethylcytidine.Citation30-Citation32 Thus, although early studies of mRNA methylation using chromatographic methods failed to identify cytidine modifications other than m5C,Citation8,Citation9 it is possible that the cytosines identified in this study bear another BS-resistant modification.
With XIST, we found that about 20% of clones showed a varying pattern of m5C, whereas in HOTAIR, C1683 was always methylated. The lower frequency of methylation in XIST is in accord with methylation frequencies observed for specific positions within tRNA.Citation16 In XIST, methylation of a fraction of the molecules may serve to functionally distinguish the methylated species from the bulk XIST RNA (e.g., by their PRC2-binding abilities). It is also possible that only one of the two RNA species that are transcribed from the XIST locus (i.e. XIST or RepA)Citation25 may be subject to methylation. However, the existence of RepA has so far only been confirmed for mouse embryonic cells. Alternatively, the lower frequency of non-converted Cs may also indicate that these positions bear a modification such as m4mC, which confers less resistance against deamination by BS treatment (Fig. S1B).Citation16
Our data together with the recent reports on the global analysis of m5C and m6ACitation12-Citation14 point to a strong potential for important biological functions of poly(A)RNA modifications. In analogy to the regulatory mechanisms acting on DNA and histones, RNA modifications might constitute what was recently dubbed “RNA epigenetics.”Citation33 The further analysis of methylation of regulatory RNAs, such as HOTAIR and XIST, may reveal new and interesting roles in various processes including transcription, translation and RNA function.
Materials and Methods
Cell lines and tissues
HEK293 (human embryonic kidney), NT2 (human neuron committed teratocarcinoma), Hs578T and BT20 (human mamma carcinoma), HOC7 (human ovarian carcinoma), female BL6 mouse embryo at developmental stage E17.
Isolation of RNA
RNA was isolated using TRizol (Sigma-Aldrich), digested with DNase I (Ambion) and purified by phenol-chloroform extraction. For eukaryotic cells, RNA was purified from isolated nuclei. To this end, cells were lysed in hypotonic buffer (10 mM HEPES-KOH, pH7.9, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, 0.5 mM PMSF) and nuclei were pelleted by centrifugation at 4,000 rpm before RNA extraction. Poly(A) RNA was prepared using the Dynabeads mRNA Purification Kit (Invitrogen).
Bisulfite sequencing
To establish the BS sequencing protocol, total RNA from E. coli was subjected to either one round of incubation for 5 min at 70°C followed by 90 min at 64°C (Fig. S1A) or three cycles of alternating incubations of 5 min at 70°C/45 min at 64°C (Fig. S1B) using the EZ RNA Methylation Kit, Beta Version (Zymo Research) according to the manufacturer’s instructions. Since the latter condition resulted in the most efficient deamination, this protocol was used for the treatment of 1–2 µg of eukaryotic poly(A)RNA. Removal of bisulfite, desulphonation and washing steps were performed by binding the RNA to spin columns provided in the kit. To determine the fragment size of BS-treated poly(A)RNA, its absorbance profile was obtained using an Agilent 2100 Bioanalyzer (Fig. S1C). The suitability of fragmented BS-treated RNA for PCR amplification was determined by performing PCR with primers recognizing deaminated sequences in GAPDH, RPS6, XIST and HOTAIR (Fig. S1D).
Reverse transcription, cloning and sequencing of BS-treated RNA
Reverse transcription of BS-treated RNA was performed with random hexamer oligonucleotides and GoScript Reverse Transcriptase (Promega). For analysis of HOTAIR, cDNA was subjected to a single round of amplification using a HOTAIR-specific forward primer together with a reverse primer bearing a random 4 nt bar code sequence in addition to an adapter sequence. PCR was performed with primers specific for deaminated sequences, and amplicons were cloned into pGEM-T Easy vector (Promega) followed by sequencing of several individual clones. Deamination rates (DR) were calculated as the percentage of non-deaminated cytosines out of all sequenced cytosines of a particular amplicon. Occurrence of a non-deaminated C at a specific position in more than 15% of clones was considered to represent methylation. All primer sequences are listed in Table S1.
In vitro transcription and RNA folding
To generate the template for in vitro transcription, the entire A-region of XIST was amplified by PCR with primers listed in Table S1. One µg of template PCR product was used for in vitro transcription using the MEGAscript Kit (Ambion) according to the manufacturer’s instructions. The RNA product was precipitated with 2.2 M LiCl, dissolved in RNase-free a.d. and digested with DNase I (Ambion). After purification using the RNeasy Mini Kit (Qiagen), the RNA was denatured at 65°C for 30 min in an Eppendorf incubator and then left in the switched-off incubator to slowly cool to room temperature in order to permit proper folding. The refolded RNA was then mixed with E. coli total RNA at a mass ratio of 1:10 and subsequently subjected to BS sequencing.
RT-qPCR
Real-time PCR was performed with an ABI Prism 7900HT Detection System (Applied Biosystems) using cDNA prepared from the indicated cell lines () and primers as well as Taqman probes as listed in Table S1. The standard curves were generated using serially diluted solutions of standard cDNA derived from the HOC-7 ovarian carcinoma cell line. TATA-binding protein (TBP) was used as the internal reference.
3D structure modeling of XIST
Tertiary structure modeling of the XIST A-region was performed using the MC-Sym program.Citation36 The inputs were ASCII scripts describing the secondary structure of the RNA published by Duszczyk et al.Citation22 for repeats 5‒8 of XIST A region, with additional base-pairings in the polyU spacer region between R7 and R8. The specific order used to build the model was the following: stem R5, spacer between 5–6 with stem-loop, stem R6, spacer between 6–7, stem R7, spacer between 7–8 with stem-loop, stem R8. Additionally, the repeats 5–8 helix was modeled separately, with the two spacer regions connected to form a closed sequence as well as the repeats 6–7 helix. C-A and G-U interactions were included for modeling and considered as non-canonical base-pairings.Citation37,Citation38 The AUCG tetra-loop of each repeat was also scripted to correspond exclusively to the NMR structure fold published by Duszczyk et al.Citation22 Although helical stacking, as well as any other desired interaction, can be “forced” by description in MC-Sym scripts, no additional interactions were provided in the MC-Sym scripts other than the base-pairings presented in Figure S4. At each position, 25% of the cyclic building blocks were tried, the backtrack was 25% of the structure and the method was probabilistic. The maximum number of structures was fixed to 30, the computation time to 72 h and the minimal difference between two structures to 2.0 Å. The energies of the structures were directly minimized on the web server until the root-mean-square gradient (GRMS) was < 5 kcal/mol/Å.
RNA-binding assay
We synthesized an m5C-modified RNA spanning repeat 8 of XIST (5′-AUC UGC UUG m5C m5C m5C AUC GGG Gm5C m5C GCG GAU) using the 2'-O-TOM-methodology for RNA solid-phase synthesisCitation39-41 and a commercially available m5C phoshoramidite building block (Glen Research). An unmodified RNA with the same sequence was obtained from IDT. Both RNAs were repurified using Vivaspin 500 columns (Sartorius),32P-labeled at the 5′-end using T4 polynucleotide kinase (New England Biolabs) and purified again on Centri-Spin 10 columns (Princeton Separations). Binding reactions were performed by incubating 4 µg of recombinant PRC2 complex (BPS Biosciences) with either the modified or unmodified labeled RNA at equimolar concentrations in binding buffer (20 mM TRIS-HCl pH 7.9, 0.1% NP-40, 10% Glycerol) for 15 min at room temperature. The PRC2-RNA complexes were purified on anti-Flag-M2-agarose (Sigma-Aldrich). The amount of bound RNA was determined by scintillation counting of input material and extensively washed beads and expressed as percent of input material. Proteins bound to the Flag-beads were eluted in 2x SDS loading buffer and subjected to electrophoresis on 8% (PRC2) or 15% (scAb) SDS polyacrylamide gels with subsequent staining by Coomassie brilliant blue.
Author note: Please add in-text citation for References Citation34 and Citation35.
Additional material
Download Zip (1.2 MB)Acknowledgments
We thank Alessia Buzzo for technical assistance and Andreas Naschberger and Klaus Scheffzek for providing reagents. We are grateful to Jim Kadonaga and Peter Loidl for discussions and comments on the manuscript. This work was supported by the Austrian Science Fund (START Y275-B12 to A.L., P21641 to R.M., M1449 to M.F.S.) and an EMBO Long-Term Fellowship to M.F.S.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Hotchkiss RD. The quantitative separation of purines, pyrimidines, and nucleosides by paper chromatography. J Biol Chem 1948; 175:315 - 32; PMID: 18873306
- Wyatt GR. Occurrence of 5-methylcytosine in nucleic acids. Nature 1950; 166:237 - 8; http://dx.doi.org/10.1038/166237b0; PMID: 15439258
- Grosjean H. Nucleic acids are not boring long polymers of only four types of nucleotides: a guided tour. In: Grosjean H, ed. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. Austin, Texas: Landes Bioscience, 2009:1-18.
- Szyf M. Epigenetics, DNA methylation, and chromatin modifying drugs. Annu Rev Pharmacol Toxicol 2009; 49:243 - 63; http://dx.doi.org/10.1146/annurev-pharmtox-061008-103102; PMID: 18851683
- Motorin Y, Helm M. RNA nucleotide methylation. Wiley Interdiscip Rev RNA 2011; 2:611 - 31; http://dx.doi.org/10.1002/wrna.79; PMID: 21823225
- Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, et al. The RNA Modification Database, RNAMDB: 2011 update. Nucleic Acids Res 2011; 39:Database issue D195 - 201; http://dx.doi.org/10.1093/nar/gkq1028; PMID: 21071406
- Motorin Y, Lyko F, Helm M. 5-methylcytosine in RNA: detection, enzymatic formation and biological functions. Nucleic Acids Res 2010; 38:1415 - 30; http://dx.doi.org/10.1093/nar/gkp1117; PMID: 20007150
- Dubin DT, Taylor RH. The methylation state of poly A-containing messenger RNA from cultured hamster cells. Nucleic Acids Res 1975; 2:1653 - 68; http://dx.doi.org/10.1093/nar/2.10.1653; PMID: 1187339
- Salditt-Georgieff M, Jelinek W, Darnell JE, Furuichi Y, Morgan M, Shatkin A. Methyl labeling of HeLa cell hnRNA: a comparison with mRNA. Cell 1976; 7:227 - 37; http://dx.doi.org/10.1016/0092-8674(76)90022-2; PMID: 954080
- Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci USA 1974; 71:3971 - 5; http://dx.doi.org/10.1073/pnas.71.10.3971; PMID: 4372599
- Adams JM, Cory S. Modified nucleosides and bizarre 5′-termini in mouse myeloma mRNA. Nature 1975; 255:28 - 33; http://dx.doi.org/10.1038/255028a0; PMID: 1128665
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012; 485:201 - 6; http://dx.doi.org/10.1038/nature11112; PMID: 22575960
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 2012; 149:1635 - 46; http://dx.doi.org/10.1016/j.cell.2012.05.003; PMID: 22608085
- Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res 2012; 40:5023 - 33; http://dx.doi.org/10.1093/nar/gks144; PMID: 22344696
- Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA 1992; 89:1827 - 31; http://dx.doi.org/10.1073/pnas.89.5.1827; PMID: 1542678
- Schaefer M, Pollex T, Hanna K, Lyko F. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res 2009; 37:e12; http://dx.doi.org/10.1093/nar/gkn954; PMID: 19059995
- Baudin-Baillieu A, Fabret C, Liang XH, Piekna-Przybylska D, Fournier MJ, Rousset JP. Nucleotide modifications in three functionally important regions of the Saccharomyces cerevisiae ribosome affect translation accuracy. Nucleic Acids Res 2009; 37:7665 - 77; http://dx.doi.org/10.1093/nar/gkp816; PMID: 19820108
- Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ, et al. Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell 2006; 21:87 - 96; http://dx.doi.org/10.1016/j.molcel.2005.10.036; PMID: 16387656
- Chernyakov I, Whipple JM, Kotelawala L, Grayhack EJ, Phizicky EM. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5′-3′ exonucleases Rat1 and Xrn1. Genes Dev 2008; 22:1369 - 80; http://dx.doi.org/10.1101/gad.1654308; PMID: 18443146
- Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, et al. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev 2010; 24:1590 - 5; http://dx.doi.org/10.1101/gad.586710; PMID: 20679393
- Chen Y, Sierzputowska-Gracz H, Guenther R, Everett K, Agris PF. 5-Methylcytidine is required for cooperative binding of Mg2+ and a conformational transition at the anticodon stem-loop of yeast phenylalanine tRNA. Biochemistry 1993; 32:10249 - 53; http://dx.doi.org/10.1021/bi00089a047; PMID: 8399153
- Duszczyk MM, Wutz A, Rybin V, Sattler M. The Xist RNA A-repeat comprises a novel AUCG tetraloop fold and a platform for multimerization. RNA 2011; 17:1973 - 82; http://dx.doi.org/10.1261/rna.2747411; PMID: 21947263
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012; 81:145 - 66; http://dx.doi.org/10.1146/annurev-biochem-051410-092902; PMID: 22663078
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010; 329:689 - 93; http://dx.doi.org/10.1126/science.1192002; PMID: 20616235
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 2008; 322:750 - 6; http://dx.doi.org/10.1126/science.1163045; PMID: 18974356
- Wutz A. Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat Rev Genet 2011; 12:542 - 53; http://dx.doi.org/10.1038/nrg3035; PMID: 21765457
- Maenner S, Blaud M, Fouillen L, Savoye A, Marchand V, Dubois A, et al. 2-D structure of the A region of Xist RNA and its implication for PRC2 association. PLoS Biol 2010; 8:e1000276; http://dx.doi.org/10.1371/journal.pbio.1000276; PMID: 20052282
- Kanhere A, Viiri K, Araújo CC, Rasaiyaah J, Bouwman RD, Whyte WA, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell 2010; 38:675 - 88; http://dx.doi.org/10.1016/j.molcel.2010.03.019; PMID: 20542000
- Kaneko S, Li G, Son J, Xu CF, Margueron R, Neubert TA, et al. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev 2010; 24:2615 - 20; http://dx.doi.org/10.1101/gad.1983810; PMID: 21123648
- Jin SG, Kadam S, Pfeifer GP. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res 2010; 38:e125; http://dx.doi.org/10.1093/nar/gkq223; PMID: 20371518
- Nestor C, Ruzov A, Meehan R, Dunican D. Enzymatic approaches and bisulfite sequencing cannot distinguish between 5-methylcytosine and 5-hydroxymethylcytosine in DNA. Biotechniques 2010; 48:317 - 9; http://dx.doi.org/10.2144/000113403; PMID: 20569209
- Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One 2010; 5:e8888; http://dx.doi.org/10.1371/journal.pone.0008888; PMID: 20126651
- He C. Grand challenge commentary: RNA epigenetics?. Nat Chem Biol 2010; 6:863 - 5; http://dx.doi.org/10.1038/nchembio.482; PMID: 21079590
- Parisien M, Major F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature 2008; 452:51 - 5; http://dx.doi.org/10.1038/nature06684; PMID: 18322526
- Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA 2001; 7:499 - 512; http://dx.doi.org/10.1017/S1355838201002515; PMID: 11345429
- Stombaugh J, Zirbel CL, Westhof E, Leontis NB. Frequency and isostericity of RNA base pairs. Nucleic Acids Res 2009; 37:2294 - 312; http://dx.doi.org/10.1093/nar/gkp011; PMID: 19240142
- Wachowius F, Höbartner C. Chemical RNA modifications for studies of RNA structure and dynamics. Chembiochem 2010; 11:469 - 80; http://dx.doi.org/10.1002/cbic.200900697; PMID: 20135663
- Micura R. Small interfering RNAs and their chemical synthesis. Angew Chem Int Ed Engl 2002; 41:2265 - 9; http://dx.doi.org/10.1002/1521-3773(20020703)41:13<2265::AID-ANIE2265>3.0.CO;2-3; PMID: 12203559
- Pitsch S, Weiss PA, Jenny L, Stutz A, Wu X. Reliable Chemical Synthesis of Oligoribonucleotides (RNA) with 2'-O-[(Triisopropylsilyl)oxy]methyl(2'-O-tom)-Protected Phosphoramidites. Helv Chim Acta 2001; 84:3773 - 95; http://dx.doi.org/10.1002/1522-2675(20011219)84:12<3773::AID-HLCA3773>3.0.CO;2-E