Abstract
Synthetic biology approaches often combine natural building blocks to generate new cellular activities. Here, we make use of two RNA elements to design a regulatory device with novel functionality. The system is based on a hammerhead ribozyme (HHR) that cleaves itself to generate a liberated ribosome-binding site and, thus, permits expression of a downstream gene. We connected a temperature-responsive RNA hairpin to the HHR and, thus, generated a temperature-controlled ribozyme that we call thermozyme. Specifically, a Salmonella RNA thermometer (RNAT) known to modulate small heat shock gene expression by temperature-controlled base-pairing and melting was fused to the ribozyme. Following an in vivo screening approach, we isolated two functional thermozymes. In vivo expression studies and in vitro structure probing experiments support a mechanism in which rising temperatures melt the thermometer structure impairing the self-cleavage reaction of the ribozyme. Since RNA cleavage is necessary to liberate the RBS, these engineered thermozymes shut off gene expression in response to a temperature increase and, thus, act in a reverse manner as the natural RNAT. Our results clearly emphasize the highly modular nature and biotechnological potential of ribozyme-based RNA thermometers.
Introduction
Gene expression is often regulated in response to variations of environmental parameters such as changing nutrient availability or altered physical conditions. Particularly in bacteria, regulatory RNAs are frequently involved in natural mechanisms of conditional gene expression.Citation1,Citation2 For example, riboswitches are RNA-based regulatory elements that are positioned in the 5′-untranslated region (5′-UTR) of the transcript they regulate.Citation3-Citation5 They are composed of an aptamer domain displaying high affinity and specificity for a cognate ligand. Usually, a second domain termed expression platform is located downstream of the aptamer domain. Upon ligand binding to the aptamer domain, a conformational change results in the formation of alternate folds in the expression platform that either change transcription termination or translation initiation.Citation6-Citation8 Following early proof of concept work,Citation9 the discovery of naturally occurring RNA-based switches of gene expression has sparked a series of studies demonstrating the construction of synthetic riboswitches.Citation10,Citation11 The incorporation of in vitro-selected aptamers into mRNAs resulted in genetic switches in bacteria as well as eukaryotic organisms.Citation12,Citation13 More specifically, we and others have designed artificial riboswitches based on a ligand-dependent regulation of a self-cleaving hammerhead ribozyme.Citation11,Citation14-Citation16 Although ligand-dependent ribozymes have been developed even before the discovery of widespread use of riboswitch mechanisms in nature,Citation17 such aptazymes have been applied in vivo for gene-regulatory purposes only after the discovery of catalytically well-behaved, fast-cleaving variants of the hammerhead ribozyme comprising stem I/II interactions.Citation18-Citation20 In our opinion, ribozyme-based devices have the advantage of almost universal applicability for controlling RNA functions. Apart from regulating mRNA translation in bacteria and mRNA integrity in mammalian cells,Citation21,Citation22 they can be utilized in order to control the activity of tRNAs,Citation23 16S rRNA,Citation24 as well as RNAi in mammalia.Citation21 In addition, they can be combined in a modular fashion in order to yield two-input Boolean logic operators.Citation25
In addition to small molecule-responsive RNA sensors, gene expression can be controlled by temperature-sensitive RNA structures, so-called RNA thermometers (RNATs).Citation26 All known RNATs modulate translation efficiency by temperature-responsive RNA structures. Most of them function in a zipper-like manner by sequestering the Shine-Dalgarno (SD, part of the ribosome-binding site) sequence at low temperatures, hence inhibiting translation initiation.Citation27 At elevated temperatures, the secondary structure unfolds and liberates the SD sequence allowing translation of the downstream gene (). Typical RNAT-controlled genes are either heat shock genesCitation28-Citation31 or virulence genes.Citation32,Citation33 FourU-type RNAT control both classes of genes.Citation32,Citation34 They are characterized by four uridines that mask the SD sequence at low temperatures. The best studied example is located in the 5′-UTR of the Salmonella enterica small heat shock gene agsA.Citation34 The second of two short hairpins, referred to as fourU hairpin in the remainder of this article, pairs with the SD sequence and regulates expression by heat-induced zipper-like melting.Citation35,Citation36 Based on the simple melting principle, artificial RNATs comprised of one or two stem-loop structures have been designed.Citation37,Citation38 Entirely unrelated elements with RNAT-like behavior are four-stranded G-rich sequences capable of forming RNA quadruplex structures in Escherichia coli. Potential quadruplex sequences were positioned in a way that formation of the quadruplex structure resulted in masking of the SD sequence. Thermal destabilization of that structure resulted in increased gene expression.Citation39
Figure 1. Design and mechanism of artificial HHR-based RNA thermometers. (A) General mechanism of naturally occurring RNA thermometers. A hairpin structure (blue) masks the Shine-Dalgarno sequence (SD; red) at low temperatures. Upon increased temperatures, melting of the secondary structure liberates the SD and translation is turned ON. 30S, small ribosomal subunit. (B) General design of a HHR-based RNA thermometer (Thermozyme). A temperature-sensing hairpin (blue) is fused via a communication module (green) to a hammerhead ribozyme with an extended stem I masking the SD (red). At low temperatures, the secondary structure permits the self-cleaving reaction of the ribozyme, liberating its SD and allowing translation of a downstream gene. A temperature increase results in melting of the thermosensing hairpin, thereby preventing self-cleavage and translation is shut OFF. Stems are indicated with roman numbers; the ribozyme cleavage site is marked by a red arrowhead. (C) Detailed thermozyme sequence. Stem III of the Schistosoma mansonii hammerhead ribozyme is exchanged against the temperature-sensing second hairpin of the Salmonella agsA thermometer (fourU hairpin). Red, SD of the downstream open reading frame; blue, temperature-sensing fourU hairpin; green, randomized nucleotides of the communication module; boxed nucleotides, ribozyme-inactivating point mutation (A→G).
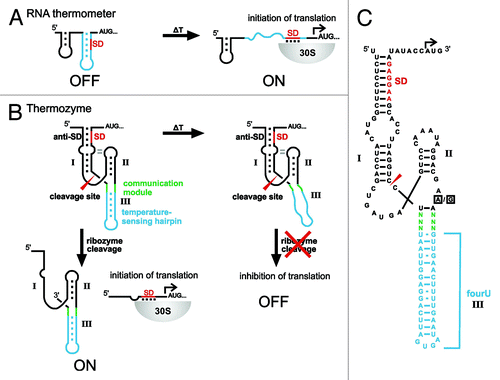
All currently known natural and synthetic RNAT control translation initiation. Therefore, we asked whether RNATs controlling other biological processes can be designed. Here, we exploit the simplicity of the Salmonella fourU hairpin to design HHR-based RNAT (thermozymes). Previously, we achieved ligand-dependency of artificial gene switches by attaching an theophylline aptamer to stem III of the ribozyme.Citation15 Screening for optimized linker sequences is often required to facilitate communication between ligand binding and ribozyme activity. In this study, we used the Salmonella fourU stem-loop as a building block to implement temperature sensitivity into an RNA-based device for artificial temperature control of gene expression. The constructed HHR-based RNATs respond to increased temperatures by an initial melting of the temperature-sensitive hairpin while the overall structure stays intact (). With our design, artificial RNA thermosensors are obtained, which show a temperature-dependency inverse to the parental, naturally occurring thermometers.
Results
Construction of Thermozymes
In order to construct a synthetic HHR-based RNAT, we attached the thermosensitive fourU hairpin to the ribozyme in place of stem III of a parental HHR derived from Schistosoma mansoni (SC-HHR), see . The position was chosen in order to preserve stem I/II interactions that facilitate fast cleavage kinetics at physiological Mg2+ concentrations in naturally occurring HHRs.Citation18-Citation20 To screen for sequences that respond to changing temperatures with altered gene expression, we randomized six nucleotides (, communication module highlighted in green) located at the junction between the ribozyme core and the fourU hairpin. A pool of different clones was constructed by inserting the randomized ribozyme sequences upstream of the eGFP gene for efficient in vivo screening. Single clones were isolated and screened for temperature responsiveness. For this purpose, the bacteria were grown at 20°C and 37°C followed by determination of gene expression by fluorescence measurements. The strain harboring the eGFP plasmid showed significant increase in gene expression at 37°C compared with growth at 20°C and the parental HHR control construct (SC-HHR) harboring the constitutively active variant of the ribozyme showed slightly increased eGFP expression at 37°C (). In the screening procedure, we identified two clones with significantly higher eGFP expression at lower temperatures as compared with 37°C (). At 20°C, the two constructs F1-HHR and B6-HHR exhibited a 3.5-fold and 3-fold induction of gene expression compared with expression at 37°C (). Normalized to the expression values of the parental SC-HHR construct, the changes of gene expression amount to 5.2-fold and 4.5-fold for clones F1-HHR and B6-HHR, respectively.
Figure 2. Characteristics of functional temperature-responsive ribozyme switches. (A) Sequence and secondary structure of the parental, constitutively active construct SC-HHR containing a stable, short stem-loop in position III of the HHR. The boxed nucleotides in the catalytic core mark the position of the inactivating point mutation (A→G). (B) Sequence of the fourU hairpin attached to stem III of the HHR, boxed square, randomized positions for the screening procedure. Inset shows screened sequences of communication modules of the two temperature-responsive clones F1-HHR and B6-HHR. (C) In vivo expression levels of various HHR constructs controlling the eGFP gene in E. coli: white bars, 20°C; black bars, 37°C. inHHRs represent catalytically inactive variants generated by the point mutation marked by boxed nucleotides in 2A.
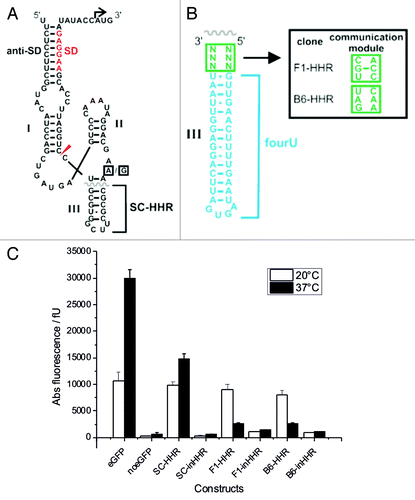
In order to validate that the observed regulation of gene expression is based on the self-cleavage reaction of the ribozymes, we introduced an inactivating A→G point mutation () in the catalytic core of the ribozyme, yielding the two inhibited constructs F1-inHHR and B6-inHHR. As expected, gene expression was impaired because translation initiation requires ribozyme activity () and was not temperature-controlled anymore (). This finding indicates that ribozyme activity is necessary for gene expression to occur.
In vitro characterization of Thermozymes
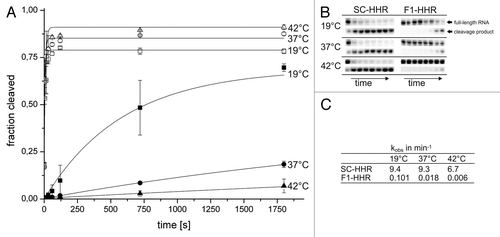
In order to characterize whether gene regulation in vivo indeed occurs via the anticipated mechanism of stem III destabilization upon increased temperature and subsequent ribozyme inactivation, we determined the temperature-dependent in vitro cleavage kinetics of the F1-HHR candidate compared with the constitutively active control ribozyme SC-HHR. The ribozyme constructs were in vitro transcribed using T7 RNA polymerase from a synthetic DNA template spanning the 5′-end of the mRNA until the end of the SD sequence, complementing stem I of the ribozyme. To prevent autocatalytic cleavage of the HHRs during transcription, we added a blocking strand at high concentrations that hybridizes to the catalytic core of the HHR and was removed during subsequent gel purification.Citation40 Ribozyme reactions were started by addition of MgCl2 to a final concentration of 0.5 mM at 19°C, 37°C and 42°C. The constitutively active SC-HHR construct containing a stable stem III shows very fast cleavage kinetics at 19°C and 37°C with only a slight decrease at 42°C (). After 5 sec, the majority of the full-length ribozyme RNA was cleaved (). For the SC-HHR variant cleavage rates of 9.4−6.7 min−1 were calculated (). These values compare well to kinetics of fast-cleaving variants at sub-millimolar Mg2+-concentrations.Citation16 In contrast to SC-HHR, cleavage of the thermozyme F1-HHR was considerably slower at all temperatures (). More importantly, the cleavage rate significantly decreased with increasing temperatures (more than 16-fold from 19°C to 42°C, see ). This temperature-dependent behavior is in accordance with the observed gene regulation in vivo.
Figure 3. Cleavage kinetics of in vitro-transcribed, purified hammerhead ribozyme variants. (A) Time course of ribozyme cleavage reactions. (SC-HHR 19°C open squares, SC-HHR 37°C open circle, SC-HHR 42°C open triangle, F1-HHR 19°C black square, F1-HHR 37°C black circle, F1-HHR 42°C black triangle). Reactions were performed using 100 nM ribozyme and were initiated with 0.5 mM MgCl2. Samples were taken at 0, 5, 15, 30, 60, 120, 720, 1,800 sec, analyzed by denaturing PAGE and band intensities were quantified. Errors have been calculated from three independent experiments. (B) PAGE-analysis of cleavage reactions. Parts of the gel corresponding to the full-length RNA and 5′ cleavage product are shown. (C) kobs values (min−1) determined for the SC-HHR and the F1-HHR at different temperatures.
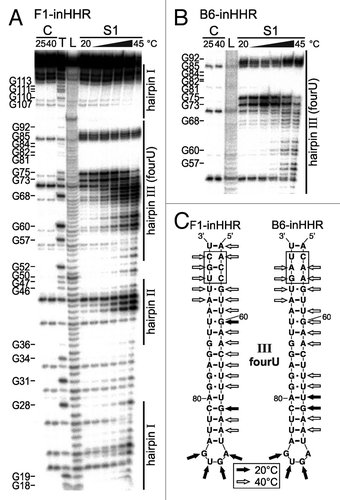
In order to demonstrate temperature-controlled melting of stem III as basis for the thermozyme action, we performed structure-probing experiments of in vitro-transcribed HHR constructs. To prevent self-cleavage of the ribozymes prior to and during the probing procedure, we analyzed the variants SC-inHHR and F1-inHHR inactivated by the A→G point mutation in the catalytic core of the ribozyme. In general, probing with nucleases S1, V1 and T1 resulted in a cleavage pattern consistent with the predicted secondary structures of the HHRs (). RNase S1 is known to cleave single-stranded regions, whereas V1 is specific for double-stranded and stacked regions. T1 preferentially cleaves 3′ of single-stranded guanines. The SC-inHHR construct containing a stable stem III structure shows basically the same cleavage pattern at both 25°C and 42°C (). For example, G residues in stem III did not become accessible to RNase T1 at 42°C. In contrast, construct F1-inHHR shows significant changes in RNase susceptibility at different temperatures (). At low temperatures the overall structure is comparable to the stucture of SC-inHHR while at higher temperatures cleavage of hairpin III resembled that of the original Salmonella agsA thermometer.Citation34 More specifically, the thermosensing fourU structure was protected from RNase S1 and T1 cleavage at 25°C, while nucleotides in this region were readily cleaved at 42°C.
Figure 4. Structural analysis of the inactive hammerhead ribozyme SC-inHHR and the inactive thermozyme variant F1-inHHR. Enzymatic cleavage of 5′ end-labeled RNA was performed with nuclease S1 (0.25 U), RNase V1 [0.002 U (A) or 0.004 U (B)] and RNase T1 (0.005 U) at 25 and 42°C. RNA fragments were separated on an 8% polyacrylamide gel. Lane C, RNA treated with water instead of RNase served as control; lane T, RNase T1 cleavage at 50°C; lane L, alkaline ladder. (A) Structure probing of the inactive ribozyme SC-inHHR (left) and secondary structure model with probing results at 42°C (right). Boxed nucleotide: position of the inactivating mutation of the HHR (A→G). Cleavage sites introduced at 42°C by nuclease S1 and RNase V1 are indicated by arrows. (B) Structure probing of the inactive thermozyme F1-inHHR (left) and secondary structure model with probing results (right). Boxed nucleotide: position of the inactivating mutation of the HHR (A→G). Cleavage sites introduced by RNase T1 and RNase V1 at 25°C are indicated by arrows. Additionally, RNase T1 cleavage sites at 42°C are marked by arrows with asterisks.
![Figure 4. Structural analysis of the inactive hammerhead ribozyme SC-inHHR and the inactive thermozyme variant F1-inHHR. Enzymatic cleavage of 5′ end-labeled RNA was performed with nuclease S1 (0.25 U), RNase V1 [0.002 U (A) or 0.004 U (B)] and RNase T1 (0.005 U) at 25 and 42°C. RNA fragments were separated on an 8% polyacrylamide gel. Lane C, RNA treated with water instead of RNase served as control; lane T, RNase T1 cleavage at 50°C; lane L, alkaline ladder. (A) Structure probing of the inactive ribozyme SC-inHHR (left) and secondary structure model with probing results at 42°C (right). Boxed nucleotide: position of the inactivating mutation of the HHR (A→G). Cleavage sites introduced at 42°C by nuclease S1 and RNase V1 are indicated by arrows. (B) Structure probing of the inactive thermozyme F1-inHHR (left) and secondary structure model with probing results (right). Boxed nucleotide: position of the inactivating mutation of the HHR (A→G). Cleavage sites introduced by RNase T1 and RNase V1 at 25°C are indicated by arrows. Additionally, RNase T1 cleavage sites at 42°C are marked by arrows with asterisks.](/cms/asset/c65e2be8-6cfc-4624-a693-36411cdf29e5/krnb_a_10924482_f0004.gif)
Fine-mapping of Thermozyme structures
In order to further characterize specific melting of the thermosensitive fourU hairpin, we performed enzymatic cleavage in a temperature range from 20−45°C. The cleavage pattern for construct F1-inHHR shows that the nucleotides of the thermosensing hairpin III are accessible for RNase S1 cleavage at higher temperature indicating melting of the hairpin (). In addition to structural changes in the RNAT module, the catalytic core and parts of stem II also showed partial melting at elevated temperatures, consistent with inactivation of ribozyme catalysis. To address whether the second thermozyme candidate B6-HHR behaves like F1-HHR, we compared the temperature-dependent structural alterations of B6-inHHR with F1-inHHR. Both showed a similar melting profile regardless of the different communication modules ().
Figure 5. Temperature-dependent alterations of the inactive thermozymes F1-inHHR and B6-inHHR. Enzymatic cleavage of 5′ end-labeled RNAs with RNase S1 (0.25 U) was performed in a temperature range from 20‒45°C in intervals of 5°C. RNA fragments were separated on an 8% polyacrylamide gel. Lane C, RNA treated with water instead of RNase at the indicated temperatures served as control; lane T, RNase T1 cleavage at 50°C; lane L, alkaline ladder. (A) Fine-mapping of the inactive thermozyme variant F1-inHHR. (B) Fine-mapping of the inactive thermozyme variant B6-inHHR. Detail of the gel corresponding to hairpin III (fourU hairpin) of the thermozyme is shown. (C) Structure model of the thermosensing hairpins III of both thermozyme variants with results of the enzymatic probing. Cleavage sites introduced by RNase S1 at 20 and 40°C are indicated by arrows. The communication modules are marked by boxes.
Discussion
Temperature-responsive HHRs were constructed by the fusion of a temperature-labile hairpin originating from a natural RNA thermometer to a self-cleaving hammerhead ribozyme. By screening for optimized sequences, two thermozymes were obtained that act as inverted, artificial RNATs when utilized as control elements for gene expression within a 5′-UTR of an E. coli mRNA. So far, aptamers binding several ligands were combined with HHRs functioning as expression platforms in order to control mRNAs, tRNAs, 16S rRNA as well as RNAi in organisms as diverse as bacteria and mammalian cells.Citation11,Citation15,Citation16,Citation21,Citation23-Citation25,Citation41
The present work shows that a physical stimulus such as a change in temperature can also be used in order to regulate gene expression via a hammerhead-mediated mRNA cleavage reaction. Importantly, most natural RNAT and all previously engineered RNAT are composed of secondary structures that mask the SD sequence.Citation37,Citation38 At elevated temperatures, gene expression is turned on by increased ribosome binding due to a more accessible, single-stranded SD region. In contrast, melting of the same thermo-sensitive structure in our artificial HHR design results in slowing down ribozyme cleavage rates, which, in turn, results in inhibition of gene expression since the HHR itself masks the SD region in the non-cleaved state. In conclusion, our design allows the construction of artificial RNA thermosensors with inversed temperature behaviors.
Modular RNA-based building blocks might be useful for future applications within the field of synthetic biology since they represent gene regulatory elements that do not require protein cofactors. Even simpler, there is also no need for externally added ligands in order to control gene expression. In addition to the potential applications of such artificial gene control devices in synthetic biology and biotechnology, the presented work demonstrates that a temperature-sensitive fourU hairpin adopted from the Salmonella agsA gene can be used as a modular component in combination with other functional RNA units. Although some other parts such as the catalytic core and stem II of the ribozyme are destabilized as well, the fourU structure melts rather specifically at increased temperatures and, thus, provides a basis for rendering the HHR temperature-responsive. The possibility to confer temperature regulation to a ribozyme both in vivo and in vitro by fusion with a natural RNAT hairpin emphasizes the remarkable modular nature of functional RNA motifs.
Materials and Methods
Plasmid construction
All constructs were introduced into a plasmid harboring the parental HHR derived from Schistosoma mansoni (SC-HHR) (HHR_pET16b_eGFP)Citation15 by PCR using Phusion DNA Polymerase (Finzyme) and sequence-specific primers with the designed construct sequences attached to the 5′ end of the primers. The following primers were used:
Pet16b_RBS_hp2_Fw: CTATTCAAAAGTTCAACNNNTTTCGTCCTATTTGGGACTCATCAGC
Pet16b_RBS_hp2_Rv: TGATTCAGGAGGTTAATNNNTCCTGGATTCCACGAAGGAGATATACC
N represents an unbiased random position generated during solid phase DNA synthesis using a 1:1:1:1 mixture of nucleoside phosphoramidites. Following the PCR, the template plasmid was digested with DpnI. The PCR products were ligated (Quick Ligase, NEB) and afterwards transformed into E. coli BL21(DE3) gold (Stratagene). Single colonies were picked and grown in LB-Medium supplemented with 100 µg ml−1 carbenecilline (Roth). To confirm successful cloning, the cloned plasmids were isolated (Miniprep Kit, Qiagen) and sequenced.
In vivo screening for functional Thermozymes
For identifying temperature-responsive HHR constructs, single clones of the transformed thermosensor-HHR pool were picked into 96 deep well plates. The expression of eGFP of out-grown cultures at room temperature and at 37°C was compared. In detail E. coli cells [BL21 (DE3), Stratagene] harboring the various plasmids were grown in Luria-Bertani (LB) medium with carbenicillin (100 µg ml−1) at 37°C under continuous shaking (1,050 r.p.m.). After 24 h, bacteria were diluted in fresh medium in a 96 deep well format and grown at room temperature under continuous shaking (1,050 r.p.m) for 2 d. After the first 24 h, 1 µl of the cell culture at room temperature was used to inoculate fresh medium in a 96 deep well format and grown at 37°C under continuous shaking (1,050 r.p.m) for 24 h, hence obtaining two cell cultures (one at room temperature and one at 37°C) after further 24 h. Two hundred µL of each culture were transferred into 96-well microplates and the fluorescence of the expressed eGFP was measured with a plate reader, using excitation wavelength of 488 nm and emission wavelength of 533 nm. The measured fluorescence was normalized to the cell optical density, measured at 600 nm.
Ribozyme kinetics
For in vitro transcription of ribozymes, a T7 RNA polymerase promoter was attached to the 5′-end of the ribozyme sequences. Synthetic DNA templates of SC-HHR and F1-HHR were PCR amplified using primers and templates shown in . The PCR products were purified by ethanol precipitation and subsequently in vitro transcribed using T7 RNA polymerase (Fermentas) in Transcription Buffer (40 mM TRIS-HCl pH 7.9, 6 mM MgCl2, 10 mM DTT, 10 mM NaCl, 2 mM spermidine, 0.5 mM ATP, 2 mM CTP, 2 mM GTP and 2 mM UTP) and 1 µCi 32P-α-ATP. One hundred µM blocking strand (ATTTGGGACTCATCAGCTGG) was added to the reaction mixture to prevent autocatalytic cleavage of the ribozymes during transcription. In vitro transcription was performed at 37°C for 2 h and subsequently purified by ethanol precipitation. The pellet was solved in 80 µL 0.5x stop buffer [40% (v/v) formamide, 25 mM EDTA pH 8.0, 0.012% (w/v) bromphenolblue]. The RNA was purified on an 8% denaturing PAGE. Full-length products were excised and passively eluted from the gel. RNA concentrations were determined photometrically.
Table 1. Primers, templates and 5‘-UTR sequences of ribozymes and thermozymes utilized in this study
For the determination of ribozyme activities, 100 nM of ribozyme in 50 mM TRIS-HCl pH 7.5, 100 mM KCl were heated to 94°C for 2 min and then slowly cooled to 19°C to fold the ribozyme. The mixtures were then incubated at 19°C, 37°C or 42°C and the cleavage reaction was started by addition of Mg2+ to a final concentration as indicated. Samples were taken at defined time points and the reactions were quenched by addition of 1x stop buffer. The reaction mixtures were separated on an 8% denaturing PAGE and visualized by phosphoimaging. To determine the cleavage rates kobs the data was fitted using the equation:
Ft = Fmax – (Fmax – F0) × e-kt
Ft, fraction cleaved at time t; Fmax, maximal fraction cleaved; F0, fraction cleaved before the reaction was started.
Enzymatic probing of Thermozyme structures
For the generation of run-off plasmids, a fragment carrying the ribozyme sequences with the T7 promoter sequence at the 5′-end and an EcoRV restriction site at the 3′-end was amplified with the primers runoff_fw (5`-AGAAATTAATACGACTCACTATAGGGATTCCCCTTTCTCCTTCGGTA-3′) and runoff_rv (5′-TTGATATCCATGGTATATCTCCTTCGTGG-3′) and cloned into the SmaI site of pUC18. SC-inHHR, F1-inHHR and B6-inHHR RNAs were synthesized in vitro by run-off transcription with T7 RNA polymerase from EcoRV-linearized plasmids. Partial digestions of 5′-32P-labeled RNA with ribonucleases T1 (Ambion), V1 (Ambion) and nuclease S1 (Fermentas, ThermoScientific) were conducted according to reference Citation34 with the exception that 1 µl of 5x TN buffer for RNase T1 (100 mM TRIS-acetate, pH 7.5; 500 mM NaCl), 1 µl of 10x RNA structure buffer for RNase V1 (provided with RNase) or 1 µl 5x reaction buffer for nuclease S1 (provided with nuclease) were used per reaction. The alkaline ladder was generated as described previously.Citation42
| Abbreviations: | ||
| HHR | = | hammerhead ribozyme |
| RNAT | = | RNA thermometer |
| UTR | = | untranslated region |
| SD | = | shine-dalgarno |
| RBS | = | ribosome-binding site |
Acknowledgments
We thank Astrid Joachimi and Cornelius Kullmann for assistance. This work was supported by a Lichtenberg Professorship awarded to J.S.H. by the VolkswagenStiftung, and by a grant of the German Research Foundation (DFG, SPP 1258: Sensory and regulatory RNAs in Prokaryotes) to F.N.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Scott WG, Martick M, Chi YI. Structure and function of regulatory RNA elements: ribozymes that regulate gene expression. Biochim Biophys Acta 2009; 1789:634 - 41; http://dx.doi.org/10.1016/j.bbagrm.2009.09.006; PMID: 19781673
- Ying SY, Chang DC, Lin SL. The microRNA (miRNA): overview of the RNA genes that modulate gene function. Mol Biotechnol 2008; 38:257 - 68; http://dx.doi.org/10.1007/s12033-007-9013-8; PMID: 17999201
- Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci USA 2004; 101:6421 - 6; http://dx.doi.org/10.1073/pnas.0308014101; PMID: 15096624
- Roth A, Breaker RR. The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem 2009; 78:305 - 34; http://dx.doi.org/10.1146/annurev.biochem.78.070507.135656; PMID: 19298181
- Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annu Rev Microbiol 2005; 59:487 - 517; http://dx.doi.org/10.1146/annurev.micro.59.030804.121336; PMID: 16153177
- Breaker RR. Prospects for riboswitch discovery and analysis. Mol Cell 2011; 43:867 - 79; http://dx.doi.org/10.1016/j.molcel.2011.08.024; PMID: 21925376
- Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 2002; 419:952 - 6; http://dx.doi.org/10.1038/nature01145; PMID: 12410317
- Winkler WC, Cohen-Chalamish S, Breaker RR. An mRNA structure that controls gene expression by binding FMN. Proc Natl Acad Sci USA 2002; 99:15908 - 13; http://dx.doi.org/10.1073/pnas.212628899; PMID: 12456892
- Werstuck G, Green MR. Controlling gene expression in living cells through small molecule-RNA interactions. Science 1998; 282:296 - 8; http://dx.doi.org/10.1126/science.282.5387.296; PMID: 9765156
- Suess B, Weigand JE. Engineered riboswitches: overview, problems and trends. RNA Biol 2008; 5:24 - 9; http://dx.doi.org/10.4161/rna.5.1.5955; PMID: 18388492
- Wieland M, Hartig JS. Artificial riboswitches: synthetic mRNA-based regulators of gene expression. Chembiochem 2008; 9:1873 - 8; http://dx.doi.org/10.1002/cbic.200800154; PMID: 18604832
- Hanson S, Berthelot K, Fink B, McCarthy JE, Suess B. Tetracycline-aptamer-mediated translational regulation in yeast. Mol Microbiol 2003; 49:1627 - 37; http://dx.doi.org/10.1046/j.1365-2958.2003.03656.x; PMID: 12950926
- Weigand JE, Sanchez M, Gunnesch EB, Zeiher S, Schroeder R, Suess B. Screening for engineered neomycin riboswitches that control translation initiation. RNA 2008; 14:89 - 97; http://dx.doi.org/10.1261/rna.772408; PMID: 18000033
- Wieland M, Hartig JS. Investigation of mRNA quadruplex formation in Escherichia coli. Nat Protoc 2009; 4:1632 - 40; http://dx.doi.org/10.1038/nprot.2009.111; PMID: 19876023
- Wieland M, Hartig JS. Improved aptazyme design and in vivo screening enable riboswitching in bacteria. Angew Chem Int Ed Engl 2008; 47:2604 - 7; http://dx.doi.org/10.1002/anie.200703700; PMID: 18270990
- Wieland M, Gfell M, Hartig JS. Expanded hammerhead ribozymes containing addressable three-way junctions. RNA 2009; 15:968 - 76; http://dx.doi.org/10.1261/rna.1220309; PMID: 19304923
- Soukup GA, Breaker RR. Engineering precision RNA molecular switches. Proc Natl Acad Sci USA 1999; 96:3584 - 9; http://dx.doi.org/10.1073/pnas.96.7.3584; PMID: 10097080
- Khvorova A, Lescoute A, Westhof E, Jayasena SD. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat Struct Biol 2003; 10:708 - 12; http://dx.doi.org/10.1038/nsb959; PMID: 12881719
- Martick M, Scott WG. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell 2006; 126:309 - 20; http://dx.doi.org/10.1016/j.cell.2006.06.036; PMID: 16859740
- De la Peña M, Gago S, Flores R. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J 2003; 22:5561 - 70; http://dx.doi.org/10.1093/emboj/cdg530; PMID: 14532128
- Ausländer S, Ketzer P, Hartig JS. A ligand-dependent hammerhead ribozyme switch for controlling mammalian gene expression. Mol Biosyst 2010; 6:807 - 14; http://dx.doi.org/10.1039/b923076a; PMID: 20567766
- Ketzer P, Haas SF, Engelhardt S, Hartig JS, Nettelbeck DM. Synthetic riboswitches for external regulation of genes transferred by replication-deficient and oncolytic adenoviruses. Nucleic Acids Res 2012; 40:e167; http://dx.doi.org/10.1093/nar/gks734; PMID: 22885302
- Berschneider B, Wieland M, Rubini M, Hartig JS. Small-molecule-dependent regulation of transfer RNA in bacteria. Angew Chem Int Ed Engl 2009; 48:7564 - 7; http://dx.doi.org/10.1002/anie.200900851; PMID: 19739151
- Wieland M, Berschneider B, Erlacher MD, Hartig JS. Aptazyme-mediated regulation of 16S ribosomal RNA. Chem Biol 2010; 17:236 - 42; http://dx.doi.org/10.1016/j.chembiol.2010.02.012; PMID: 20338515
- Klauser B, Saragliadis A, Ausländer S, Wieland M, Berthold MR, Hartig JS. Post-transcriptional Boolean computation by combining aptazymes controlling mRNA translation initiation and tRNA activation. Mol Biosyst 2012; 8:2242 - 8; http://dx.doi.org/10.1039/c2mb25091h; PMID: 22777205
- Storz G. An RNA thermometer. Genes Dev 1999; 13:633 - 6; http://dx.doi.org/10.1101/gad.13.6.633; PMID: 10090718
- Kortmann J, Narberhaus F. Bacterial RNA thermometers: molecular zippers and switches. Nat Rev Microbiol 2012; 10:255 - 65; http://dx.doi.org/10.1038/nrmicro2730; PMID: 22421878
- Kortmann J, Sczodrok S, Rinnenthal J, Schwalbe H, Narberhaus F. Translation on demand by a simple RNA-based thermosensor. Nucleic Acids Res 2011; 39:2855 - 68; http://dx.doi.org/10.1093/nar/gkq1252; PMID: 21131278
- Morita MT, Tanaka Y, Kodama TS, Kyogoku Y, Yanagi H, Yura T. Translational induction of heat shock transcription factor sigma32: evidence for a built-in RNA thermosensor. Genes Dev 1999; 13:655 - 65; http://dx.doi.org/10.1101/gad.13.6.655; PMID: 10090722
- Nocker A, Hausherr T, Balsiger S, Krstulovic NP, Hennecke H, Narberhaus F. A mRNA-based thermosensor controls expression of rhizobial heat shock genes. Nucleic Acids Res 2001; 29:4800 - 7; http://dx.doi.org/10.1093/nar/29.23.4800; PMID: 11726689
- Waldminghaus T, Fippinger A, Alfsmann J, Narberhaus F. RNA thermometers are common in alpha- and gamma-proteobacteria. Biol Chem 2005; 386:1279 - 86; http://dx.doi.org/10.1515/BC.2005.145; PMID: 16336122
- Böhme K, Steinmann R, Kortmann J, Seekircher S, Heroven AK, Berger E, et al. Concerted actions of a thermo-labile regulator and a unique intergenic RNA thermosensor control Yersinia virulence. PLoS Pathog 2012; 8:e1002518; http://dx.doi.org/10.1371/journal.ppat.1002518; PMID: 22359501
- Johansson J, Mandin P, Renzoni A, Chiaruttini C, Springer M, Cossart P. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 2002; 110:551 - 61; http://dx.doi.org/10.1016/S0092-8674(02)00905-4; PMID: 12230973
- Waldminghaus T, Heidrich N, Brantl S, Narberhaus F. FourU: a novel type of RNA thermometer in Salmonella. Mol Microbiol 2007; 65:413 - 24; http://dx.doi.org/10.1111/j.1365-2958.2007.05794.x; PMID: 17630972
- Rinnenthal J, Klinkert B, Narberhaus F, Schwalbe H. Direct observation of the temperature-induced melting process of the Salmonella fourU RNA thermometer at base-pair resolution. Nucleic Acids Res 2010; 38:3834 - 47; http://dx.doi.org/10.1093/nar/gkq124; PMID: 20211842
- Rinnenthal J, Klinkert B, Narberhaus F, Schwalbe H. Modulation of the stability of the Salmonella fourU-type RNA thermometer. Nucleic Acids Res 2011; 39:8258 - 70; http://dx.doi.org/10.1093/nar/gkr314; PMID: 21727085
- Neupert J, Karcher D, Bock R. Design of simple synthetic RNA thermometers for temperature-controlled gene expression in Escherichia coli. Nucleic Acids Res 2008; 36:e124; http://dx.doi.org/10.1093/nar/gkn545; PMID: 18753148
- Waldminghaus T, Kortmann J, Gesing S, Narberhaus F. Generation of synthetic RNA-based thermosensors. Biol Chem 2008; 389:1319 - 26; http://dx.doi.org/10.1515/BC.2008.150; PMID: 18713019
- Wieland M, Hartig JS. RNA quadruplex-based modulation of gene expression. Chem Biol 2007; 14:757 - 63; http://dx.doi.org/10.1016/j.chembiol.2007.06.005; PMID: 17656312
- Salehi-Ashtiani K, Szostak JW. In vitro evolution suggests multiple origins for the hammerhead ribozyme. Nature 2001; 414:82 - 4; http://dx.doi.org/10.1038/35102081; PMID: 11689947
- Kumar D, An CI, Yokobayashi Y. Conditional RNA interference mediated by allosteric ribozyme. J Am Chem Soc 2009; 131:13906 - 7; http://dx.doi.org/10.1021/ja905596t; PMID: 19788322
- Brantl S, Wagner EG. Antisense RNA-mediated transcriptional attenuation occurs faster than stable antisense/target RNA pairing: an in vitro study of plasmid pIP501. EMBO J 1994; 13:3599 - 607; PMID: 7520390