Abstract
Cross-kingdom delivery of specific microRNAs to recipient organisms via food ingestion has been reported recently. However, it is unclear if such delivery of microRNAs occurs frequently in animal organisms after typical dietary intake. We found substantial levels of specific microRNAs in diets commonly consumed orally by humans, mice, and honey bees. Yet, after ingestion of fruit replete with plant microRNAs (MIR156a, MIR159a, and MIR169a), a cohort of healthy athletes did not carry detectable plasma levels of those molecules. Similarly, despite consumption of a diet with animal fat replete in endogenous miR-21, negligible expression of miR-21 in plasma or organ tissue was observed in miR-21 −/− recipient mice. Correspondingly, when fed vegetarian diets containing the above plant microRNAs, wild-type recipient mice expressed insignificant levels of these microRNAs. Finally, despite oral uptake of pollen containing these plant microRNAs, negligible delivery of these molecules was observed in recipient honeybees. Therefore, we conclude that horizontal delivery of microRNAs via typical dietary ingestion is neither a robust nor a frequent mechanism to maintain steady-state microRNA levels in a variety of model animal organisms, thus defining the biological limits of these molecules in vivo.
Introduction
MicroRNAs (miRNA) are small, evolutionarily conserved, non-protein coding RNA molecules that negatively regulate gene expression throughout the plant and animal kingdoms. They bind specific sequences in target mRNA transcripts in order to repress translation and/or induce mRNA degradation. In animals with a circulatory system, miRNAs are also stably present in the bloodstream, and some can be directly taken up by recipient cells for control of gene expression and cellular functions important in certain diseases,Citation1-Citation5 such as modulation of atherosclerotic lesionsCitation6 and tumor metastasisCitation7 in mice. These extracellular forms of miRNAs are encapsulated through several mechanisms including packaging in phospholipid bilayer-encapsulated vesicles or exosomesCitation8 or via the formation of miRNA-protein complexes such as those bound to the Argonaute 2 protein (AGO2)Citation9 or high density lipoprotein.Citation4 These miRNAs are surprisingly stable and are resistant to degradation even in harsh environmental conditions such as acidic pH and substantial heat exposure.Citation10 MiRNAs are handled similarly in insect cells and are involved in a broad range of organismal processes.Citation11,Citation12 Mechanisms of uptake by insect cells have been well characterized and appear to involve specific double-stranded RNA (dsRNA) transporters and endocytic processes.Citation13,Citation14 Although less is known about transfer of extracellular miRNAs in insects, it has been reported that miRNAs or dsRNAs may be shared in a common pool among cells and tissues in these organisms.Citation15-Citation17
Recently, cross-kingdom transfer of plant miRNAs to mammalian organisms has been reported via oral ingestion, leading to uptake into circulating plasma, delivery to recipient tissue and consequent post-transcriptional regulation of mammalian gene expression. Specifically, two miRNAs expressed in rice, MIR156a and MIR168a, were reported to be expressed in the plasma of a Chinese cohort of individuals as well as in the plasma and organ tissue of mice.Citation18 Furthermore, MIR168a was found to directly repress expression of a mammalian mRNA target LDLRAP1 in liver and, thus, influence cholesterol regulation. Stemming from these studies, it has been debated whether animal and plant species may communicate and influence each other at the gene expression level via transfer of diet-derived miRNAs.Citation19-Citation26 If robust, cross-kingdom regulation of gene expression mediated through miRNA ingestion could revolutionize our understanding of communication in ecological networks, both natural and agricultural. As such, this paradigm has also heightened public scrutiny of artificial alterations in the human diet. The putative dietary transfer of miRNAs has been especially emphasized in relation to the advent of genetically engineered plants and animals and the unknown consequences of human intake of such diets. These issues may have broader impacts for understanding the influence of natural and genetically altered diets on all organisms. As an example, substantial interest currently exists in utilizing dsRNAs and potentially miRNAs expressed in plants to control insect pestsCitation13,Citation27,Citation28 and to influence the functions of beneficial insects, such as the honey bee. Yet, crucial to these arguments is the still unanswered question of how prevalent and how effective is this mechanism of uptake of endogenous miRNAs in various organisms consuming a normal diet. Thus, in this study, we demonstrate that horizontal delivery of miRNAs via oral ingestion of a typical diet is neither a frequent nor a prevalent event across multiple recipient animal organisms.
Results
Endogenous miRNAs are present at substantial levels in diets consumed orally by humans, mice and honey bees
We aimed to determine whether intact miRNAs are present in diets typically ingested by a variety of higher order species in the animal kingdom—humans, mice and honey bees. To that end, we selected for our study three miRNAs (MIR156a, MIR159a and MIR169a) that are conserved and highly expressed throughout the plant kingdom.Citation29 In studies of delivery of animal-based miRNAs, we chose to focus on a conserved mammalian miRNA (miR-21) given its ubiquitous and high level of expression throughout mammalian tissueCitation30 and in the circulating bloodstream.Citation31 After RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-PCR), MIR156a, MIR159a and MIR169a were detected at substantial levels in fruits common in the human diet such as ripened avocado, apple, banana, orange and cantaloupe but not in meat products, such as ham, common in a human Western diet (). Notably, the copy numbers of MIR156a and MIR159a were found to be especially elevated in many fruit and approximated the highest level of expression of any miRNA confirmed independently in other fleshy fruits such as grapes.Citation32 Conversely, the conserved animal miRNA (miR-21) was expressed at substantial levels in meat but not fruits ().
Figure 1. Substantial levels of miRNAs in oral diets consumed by humans, mice, and honey bees. (A) MiRNAs conserved in the plant kingdom were present in substantial quantity in ripened fruits, while miR-21, a conserved miRNA in the animal kingdom, was present in a common dietary source of meat (ham). & denotes that miR-21 was not detectable in fruit products, and this absence of signal was significantly different (p < 0.05) than copy number of MIR156a, MIR159a or MIR169a in a given diet, based on ANOVA and post-hoc testing. # denotes that plant miRNAs were not detectable in ham, and this absence of signal was significantly different (p < 0.05) than copy number of miR-21. (B) Conserved plant miRNAs were expressed at high levels in a custom vegetarian diet and a custom diet with soy, while miR-21 was present in a custom diet carrying animal lard and casein. & denotes that miR-21 was not detectable in the vegetarian or soy diets, and this absence of signal was significantly different (p < 0.05) than copy number of MIR156a, MIR159a or MIR169a in a given diet. # denotes that MIR-159a and MIR169a were not detectable in the casein and lard diet, and * denotes that copy number of miR-21 was significantly different (p < 0.05) than copy number of MIR156a, MIR159a and MIR169a in that diet. (C) Conserved plant miRNAs were present in substantial quantity in pollen, a primary food source for the honey bee, and in honey stored by honey bees for later consumption. In all panels, each food source was sampled and analyzed three independent times (n = 3), and error bars, which are small but present on each column, reflect SEM.
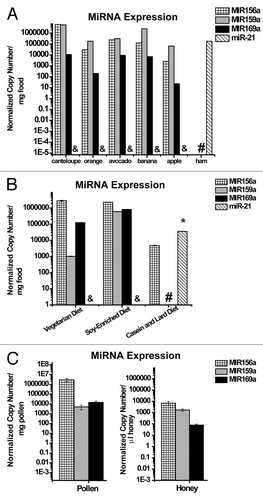
Based on a prior report that normal chow diets used to feed laboratory mice carry relatively lower levels of plant-derived miRNAs such as MIR156a,Citation18 we chose to utilize custom rodent diets enriched for plant material and thus optimized to deliver maximal levels of endogenous plant miRNAs to recipient mice. We found that a custom vegetarian mouse diet and a diet enriched in soy () both carried high quantities of MIR156a, MIR159a and MIR169a but not miR-21 (). Importantly, both of these diets carried especially elevated levels of MIR156a, which approximated the amount of MIR156a and MIR168a in rice that previously was reported to be delivered into the circulating bloodstream and organ tissue of recipient mice ingesting rice.Citation18 On the other hand, mouse diets enriched in animal products such as lard and casein purified from cow’s milk () carried elevated levels of miR-21 (). Finally, as the primary food source of honey bees, pollen and honey samples were acquired from the frames of managed honey bee hives (). MIR156a, MIR159a and MIR169a were all expressed, with MIR156a particularly elevated in pollen, in agreement with a prior report of miRNA levels in fresh pollen.Citation33 Consequently, miRNAs conserved in both plant and animal kingdoms are present in oral diets of various animal organisms and, in many cases, these miRNAs are present at especially elevated levels.
Table 1. Composition of custom mouse diets
Selected plant miRNAs are not detectable in the plasma from healthy human subjects after intake of a western diet containing fruits
The dietary records of 10 healthy collegiate varsity athletes (crew) were reviewed, and breakfast intake was documented on the day prior to plasma harvest (). While some heterogeneity of dietary intake was documented, seven of the 10 subjects reported routine intake of fruits replete in MIR156a, MIR159a and MIR169a. Consistent with previously measured copy numbers from healthy human subjects,Citation34 the extracellular, circulating form of a representative endogenous human miRNA, miR-16, was present at high levels in plasma (). In contrast, the above plant-derived miRNAs were undetectable in those same samples (). Thus, these data suggest an inability of humans to maintain steady-state levels of plant miRNAs in the circulating bloodstream after intake of a typical Western diet containing substantial plant material.
Table 2. Dietary intake prior to plasma harvest in a cohort of healthy male athletes
Table 3. MIR156a, MIR159a and MIR169a were not detectable in the plasma from ten healthy male athletes
Negligible expression of miR-21 in plasma or organ tissue in miR-21 −/− mice after oral diets replete with endogenous miR-21
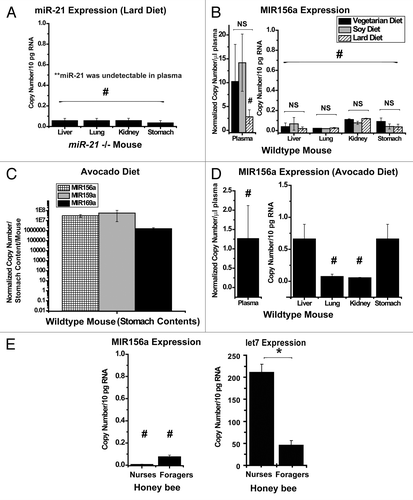
To define definitively whether intake of diets replete with endogenous mammalian miRNAs maintains detectable miRNA levels in mice, we chose to determine the presence of diet-delivered miR-21 in miR-21-null (miR-21 −/−) mice. As we have previously reported,Citation30 wild-type mice expressed high levels of miR-21 in plasma and across disparate organ tissues, while miR-21 −/− mice fed a normal chow diet expressed negligible levels of miR-21 throughout all plasma and tissue compartments. Over a 4 wk period, adult miR-21 −/− mice were fed a custom animal lard diet replete in miR-21 (27.5 ± 1.7 g, mean ± SEM, animal lard diet consumed per mouse per week, ). Afterward, plasma and organ tissue were harvested for miRNA quantification. Despite consistent plasma levels of endogenous miR-16 (Fig. S1A, left graph), miR-21 was not detectable in plasma from miR-21 −/− mice. In comparison, miR-21 was present in relatively abundant quantity in the plasma of wild-type mice (miR-21 +/+) fed a comparable diet (Fig. S1A, right graph). Furthermore, in miR-21 −/− mice, despite relatively abundant tissue levels of the ubiquitously expressed miR-195 across organs (Fig. S1A, middle graph), miR-21 was either not present or detectable at exceedingly low levels (), corresponding to less than one copy of miR-21 per cell (conservatively assuming that the typical mammalian cell expresses roughly 10 pg of total RNACitation35). Consequently, these data demonstrate that transfer of diet-derived miRNA is an extremely ineffective mechanism to augment steady-state expression of miR-21 in mice that cannot endogenously produce this miRNA.
Figure 2. Negligible steady-state expression of diet-derived miRNAs in plasma or organ tissue. (A) Oral ingestion of a lard diet replete with miR-21 did not lead to a steady-state increase of miR-21 in plasma or organ tissue in miR-21 −/− recipient mice (n = six mice per group). (B) In spite of vegetarian diets (vegetarian, black bar; soy, gray bar) containing endogenous MIR156a, MIR159a and MIR169a as compared with a diet with lower levels of plant miRNAs (lard, hatched bar), minimal expression of MIR156a in plasma was observed (left graph) with negligible steady-state expression in organ tissue (right graph) of wild-type recipient mice (n = five mice per group). Notably, in these preparations, MIR159a and MIR169a were not detectable. (C) Unprocessed avocado ingestion increases the expression of conserved plant miRNAs in the stomach contents of wild-type mice. (D) Ingestion of fresh avocado does not result in substantial steady-state expression of MIR156a in plasma (left graph) or organs (right graph) in wild-type recipient mice (n = four mice). Again, in these preparations, MIR159a and MIR169a were not detectable. (E) (Right graph) The honey bee miRNA let7 was expressed in the abdomen of both nurses and foragers. Higher levels of let7 were observed incidentally in nurses as compared with foragers. (Left graph) In spite of oral uptake of pollen and honey/nectar replete with MIR156a, MIR159a and MIR169a, negligible levels of MIR156a were detected in the abdominal tissue of recipient honey bees (n = three nurses; n = four foragers). MIR159a and MIR169a were undetectable in these tissues. In all panels, error bars reflect SEM; * signifies p < 0.05; NS signifies p ≥ 0.05. # denotes that the expression of a given miRNA was either undetectable or at the level of minimal detection. For plasma, normalized copy number is expressed per μl plasma. For animal organs, copy number is expressed per 10 pg of total RNA. For stomach contents, normalized copy number is expressed per total gastric material harvested per mouse.
Negligible expression of MIR156a, MIR159a and MIR169a in plasma or organ tissue in mice after ingestion of diets replete with these miRNAs
Correlating with the above analyses of miR-21, we aimed to determine whether oral diets containing plant miRNAs facilitate delivery and expression of these miRNAs in recipient mice. Over a 4 wk period, adult wild-type mice were fed one of three custom diets—a vegetarian diet, a soy-enriched diet or a diet replete in animal lard (). Comparable consumption of each diet was observed (25.6 ± 1.2 g, vegetarian diet consumed per mouse per week; 28.5 ± 2.1 g, soy diet consumed per mouse per week; 28.4 ± 2.2 g, animal lard diet consumed per mouse per week). Consistent plasma levels of endogenous miR-16 and consistent tissue levels of miR-195 across diets were again observed (Fig. S1B). In that context, there was a statistically non-significant trend toward increased plasma levels of MIR156a after consuming either a vegetarian or soy diet as compared with animal lard (, left graph). Notably, such levels were exceedingly low. Correspondingly, MIR156a was detectable in organ tissue but at levels corresponding to far less than one copy of miRNA per cell (, right graph) and without any evidence of increased expression after vegetarian or soy diet intake. Furthermore, regardless of diet, MIR159a and MIR169a were not detectable in either plasma or organ tissue.
Since it was possible that prior processing of these rodent diets may have affected the efficiency of putative transfer after ingestion, mice were fed unprocessed avocado (average intake of 4.5 ± 0.5 g per mouse over 24 h), a food source containing all three specific plant miRNAs (). Importantly, such intake of avocado approximated the level of intake of both MIR156a and MIR168a in rice that previously was reported to facilitate miRNA transfer to recipient mice after ingestion.Citation18 After 24 h of avocado diet, endogenous miRNAs in the plasma (miR-16) and in tissues (miR-195) were confirmed (Fig. S1C). In addition, avocado intake was further verified by documentation of substantial expression of all three plant miRNAs in stomach contents from these mice (). Despite ingestion of these miRNAs, MIR156a was measured at exceedingly low levels in both plasma (, left graph) and organ tissue (, right graph). Furthermore, neither MIR159a nor MIR169a was detected in plasma and organ tissue after avocado intake. Thus, after being fed with either processed or non-processed vegetarian diets, mice did not display evidence of substantial steady-state expression of these plant miRNAs.
Negligible expression of plant-derived miRNAs in recipient honey bee tissues
Finally, to determine whether oral consumption facilitates expression of diet-derived miRNAs in animals other than mice and humans, we extended our study to honey bees that naturally ingest pollen, nectar and processed forms of these plant products, from multiple plant species. To minimize the confounding possibility of detecting pollen-based miRNAs that had been ingested but not transferred to recipient honey bees, gut tissue was removed from nurses and foragers at the time of tissue harvest prior to RNA extraction. A near equivalence in expression of the housekeeping gene β-actin was found among these two groups (Fig. S1D). Furthermore, demonstrating that endogenous miRNA molecules were detectable in these tissue preparations, a relative abundance of the honey bee miRNA let7 was observed in both nurses and foragers (, right graph, incidentally also revealing increased expression in nurses compared with foragers). Yet, despite expression of MIR156a, MIR159a and MIR169a in pollen (), only MIR156a, but not MIR159a or MIR169a, was detected in abdominal tissue derived from nurses and foragers, but again at exceptionally low levels (, left graph). Thus, mirroring our findings in mice and humans, a typical diet of pollen and nectar is ineffective at increasing steady-state diet-derived miRNA levels in honey bees.
Discussion
Here, we have found that a number of higher order animal organisms, including honey bees, mice and humans, routinely ingest substantial quantities of plant-specific and animal-specific miRNAs. In spite of such ingestion, we find little evidence of significant steady-state expression of those miRNAs in recipient organisms (< 1 copy per cell in various organ tissues). Furthermore, based on prior observations that poorly expressed miRNAs (< 100 copies per cell) do not repress target-containing transcripts,Citation36 it is unlikely that dietary miRNAs are routinely spread to plasma or organ tissue in quantities that are adequate for pervasive engagement of direct mRNA targets. These results dampen our enthusiasm regarding the widespread potential for miRNA-based cross-kingdom modulation of gene expression, at least via normal dietary intake and through canonical modes of action of miRNAs.
Since the first reports of inhibitory RNA nearly 20 y ago, achieving efficient RNA delivery in mammalian organisms through an oral route has been challenging. Thus, perhaps it is not surprising that even endogenously packaged forms of miRNAs, although quite stable in the circulating mammalian blood stream and in various ex vivo contexts (e.g., in food sources, ), may nonetheless also be prone to poor uptake and delivery after ingestion. More recent methods to engineer specific packaging of RNA particles (i.e., nanoparticles) to withstand the harsh conditions of the gastrointestinal tract have shown promiseCitation37 and appear more likely to offer viable options for miRNA delivery in mammalian species.
Furthermore, beyond invoking specific deficiencies in the mechanism of delivery of miRNAs through the mammalian digestive tract per se, it is conceivable that the typical mammalian diet simply contains insufficient quantities of miRNAs for biological activity across multiple tissue compartments. This argument is consistent with our observations that, on average, when MIR156a is expressed at higher levels in food as compared with MIR159a and MIR169a, it is more easily detected in recipient organisms, albeit at extremely low levels (). Thus, improvement in delivery may be theoretically possible with increased ingestion, but these dietary alterations would be challenging under normal circumstances. For example, assuming that the average human body contains 1013 cellsCitation38 and that each miRNA on average must reach at least 100 copies per cell to induce canonical target gene repression,Citation36 at least 1015 copies of a given plant miRNA must be ingested and delivered for widespread biological activity. Based on our findings that ripened cantaloupe contains approximately 6 × 106 copies of MIR156a per mg of fruit, an individual would need to consume 1,670 kg of cantaloupe just to realize this copy number in the gastrointestinal tract, and substantially greater consumption would be necessary to overcome inefficiencies of transfer to the rest of the body. Although miRNA copy number can change in fruit during ripening,Citation39,Citation40 the degree of known natural variations would be insufficient to make this level of delivery plausible. Furthermore, based on recent estimates of fruit and vegetable intake in Europeans ranging from 103−454 g per day,Citation41 clearly this amount of consumption is not attainable in a reasonable amount of time by typical dietary standards.
On the other hand, Zhang and colleagues have proposed that improvement in delivery is possible with increased ingestion, as reflected by their findings of increased MIR168a delivery in mice after a rice diet as compared with normal chow diet that carries lower levels of that miRNA.Citation18 Importantly, however, the extent of miRNA delivery reported by Zhang and colleagues is not consistent with our results. Specifically, although Zhang and colleagues reported a relative abundance of MIR156a in human sera (approximately 10% of the levels of endogenous miR-16), we found that plasma expression is undetectable in a cohort of healthy athletes (), despite documented ingestion of fruit carrying concentrations of MIR156a (~6 × 109 copies per g canteloupe) () comparable to those concentrations found in rice (5.72 fmol/g = 3 × 109 MIR156a copies per g rice).Citation18 Our findings are further supported by independent analyses of plant miRNA sequences in larger cohorts of human sera.Citation19 Furthermore, although Zhang and colleagues found a relatively high level of plant-derived miRNAs in plasma and organ tissue in mice after rice ingestion, we found negligible spread of multiple plant-derived miRNAs when using diets comparably replete in MIR156a-processed vegetarian/soy diets (i.e., ~1 × 1010 MIR156a copies ingested after 24 h) or avocado diet (i.e., ~1.1 × 109 MIR156a copies ingested after 24 h). To explain these discrepancies, we do not rule out the possibility of specialized circumstances that could facilitate transfer and perhaps even target miRNA to particular tissue niches after oral ingestion. For example, miRNAs specially packaged in breast milkCitation42,Citation43 have been reported with immune modulatory featuresCitation44,Citation45 and could influence intestinal immune function in the recipient infant. However, in total, our findings unambiguously demonstrate that the spread of diet-derived miRNA to higher organisms is not ubiquitous and in fact is limited at best. Consequently, our findings may temper the more publicized worries regarding abnormal miRNA transfer and widespread activity after genetic manipulation of food products.
Alternatively, in some invertebrates, oral intake of extracellular RNA has proven to be a more efficient method for delivery into recipient tissue. Specifically, uptake of dsRNAs after ingestion has been reported in multiple insect taxa,Citation13 and delivery of miRNAs by ingestion specifically has been effective in C. elegans.Citation46 In some cases, targeting of insect gene expression through ingestion of dsRNA hairpins transgenically expressed in plant tissue has also been achieved. However, the efficiency of uptake is highly variable in a species-dependent manner.Citation47 Importantly, in the honey bee, high dose injection of dsRNAs into the hemocoel has been used for delivery and inhibition of target genes in multiple tissues.Citation17,Citation48 Some studies have further indicated that honey bees represent a species for which ingestion of dsRNAs can produce marked knockdown of target genes in the digestive tractCitation49-Citation55 and other tissues at distal sites.Citation51,Citation56 In fact, direct cross-species transfer of dsRNAs is possible in honey bees, where the mite V. destructor can take up dsRNAs from one bee and pass it to a recipient via its feeding mechanism.Citation56 Yet, despite these reports regarding exogenous dsRNA delivery, our findings clearly demonstrate the absence of plant-derived miRNAs in recipient honey bees in the course of the normal pattern of pollen and honey/nectar ingestion. These discrepancies could reflect differences in either uptake or processing mechanisms of dsRNAs vs. miRNAs in the insect.Citation57 Alternatively, similar to the mammalian system, transfer of endogenously consumed plant miRNAs may be limited by insufficient copy number in pollen, as opposed to high concentrations that can be delivered orally with exogenously supplied dsRNAs. Using estimates of the number of cells in an adult bee (107, see Materials and Methods for details); the rate of pollen (0.2–0.4 mg per dayCitation58) and honey or nectar consumption (50–160 mg honey or 100–320 mg of nectar per dayCitation59) and miRNA copy number in both pollen and honey (), we calculate that an adult honey bee consumes approximately 5 × 106 copies of MIR156a per day, thus allowing for delivery 0.5 three copies per cell if 100% transfer of the diet-derived miRNA were achieved. Consequently, typical dietary intake of honey bees may be too little for more global miRNA delivery to the entire adult organism; and this possibility is consistent with both our findings () and independently published results studying other insects feeding on plants.Citation19 In contrast, a recent study has indicated that insect-derived miRNAs may be transferred to larval honey bees via inoculation of royal jelly with miRNAs by adult nurse bees, leading to specific effects on gene expression during larval development.Citation60 Given that oral dsRNA delivery can vary depending on insect life-stage,Citation13 it is possible that specific circumstances in the larvae could enhance accumulation of these miRNAs in recipient tissues. Nonetheless, by identifying a virtual absence of pollen-based miRNAs in adult honey bees ingesting a typical dietary load, our results greatly clarify the ecological limits of any natural miRNA-based communication between the adult bee and its primary food source.
In that context, is the presence of intact miRNAs in dietary food sources otherwise without meaningful biological function? Notably, our results suggest that low levels of orally ingested miRNAs at times are detectable but at concentrations that would make canonical functions of miRNAs implausible.Citation36 Nonetheless, although not yet proven, alternative mechanisms can be envisioned by which low level delivery of diet-derived miRNAs could carry physiologic significance. First, it is conceivable that specific tissue niches can act as depots of orally ingested miRNAs, thus effectively concentrating miRNAs to biologically meaningful levels for target gene regulation. Alternatively, delivery of ingested miRNAs may allow for non-canonical functions beyond direct intracellular target engagement such as engagement of cell-surface receptors that promote intracellular signaling cascades, as previously described in cancer cells.Citation61 Finally, given their anatomic proximity, oral ingestion of miRNAs in mammals could be envisioned as a means to facilitate more efficient delivery to bacteria colonizing the gastrointestinal tract, thus allowing for cross-kingdom communication from diets derived from higher order organisms (such as plants and animals) to prokaryotes.
Yet, beyond the concept of oral miRNA delivery, given the increasing appreciation of the delivery and actions of plasma-based miRNAs in vivo, we can envision more likely scenarios of horizontal transfer of miRNAs in humans that bypass the digestive tract altogether. Most notably, the possibility of miRNA delivery to recipient individuals after blood transfusion and solid organ transplantation has yet to be explored adequately. Especially in the setting of solid organ xenotransplantation, cross-species miRNA transfer with substantial biological consequence remains plausible. Furthermore, alternative endogenous mechanisms may exist that facilitate delivery of miRNAs for mucosal uptake independent of gastric transit, including anal or vaginal intercourse or the increasingly utilized procedure of fecal transplantation for treatment of C. difficile infections.Citation62 However, these possibilities await definitive confirmation.
In summary, the impact of miRNA biology continues to be wide reaching and often surprising. Nonetheless, in multiple model animal organisms, we describe here a near absence of diet-derived miRNAs in recipient tissue despite the presence of both plant and animal miRNAs in their food sources. Consequently, we believe that our data are important in defining the limits of the biology of miRNA and will guide the focus of future studies to concentrate on the more robust functions of these molecules in vivo.
Materials and Methods
Animals and diets
The generation of mmu-miR-21 −/− mice was previously described,Citation63 and these mice were a generous gift from S. Orkin (Boston Children’s Hospital). Otherwise, C57Bl6 wild-type male littermate mice were used (8–12 wk of age) for these experiments (Taconic). The Harvard Center for Comparative Medicine approved the use of animals. Custom diets were purchased from Harlan Laboratories: vegetarian diet (TD.2016); soy-Enriched diet (TD.97053) and casein and lard diet (TD.06414) ().
Honey bees were collected from outbred colonies in Williamstown, consisting of a typical mix of A. mellifera subspecies found in North America, pollen and honey were taken from these same hives. Only visibly healthy bees were collected, and all source colonies were visually inspected for symptoms of common bacterial, fungal and viral diseases of honey bees. The average number of cells in a honey bee was calculated using the following equation: Number of cells in a honey bee = mass of an average worker bee (90 mg)/mass of an individual cell. Mass of an individual cell was calculated using the equation: mass = cell volume × cell density where an average cell volume is estimated at 10 μm3 with a density of 1. Importantly, use of this same calculation resulted in the correct estimation of cells in the human body (1013 cells).Citation38
Human participants
Written informed consent was obtained from all participants; this conformed to the standards set by the latest revision of the Declaration of Helsinki. Before study initiation, the Partners Human Research Committee approved all protocols for human plasma study and collection. As chosen from a previously published cohort,Citation31 male collegiate crew athletes from Harvard University were chosen for study without significant medical problems and taking no prescription or over-the-counter medications.
Plasma collection from humans and mice
From humans, venous blood was collected in standard vacutainer tubes with K+-EDTA anticoagulant (). To do so, a 20-gauge intravenous catheter was placed antiseptically into a dorsal hand vein or a vein in the distal forearm as dictated by favorable anatomy using the Seldinger technique and then secured with tape. The IV catheter flushed was flushed with sterile saline to help maintain patency after each blood draw. Plasma was extracted after standard centrifugation of blood, followed by storage at -80°C. From mice, venous blood was collected into EDTA anticoagulant by syringe withdrawal directly after right ventricular puncture, followed by plasma extraction as previously described.Citation30
Plasma RNA extraction
RNA extraction from plasma was previously described.Citation31 Briefly, thawed plasma samples were centrifuged (13,000 rpm × 10 min), and plasma supernatant was aliquoted for storage at -80°C or further analysis. Specifically, to a standard volume of each plasma supernatant (100 μl), 2 ficomoles were added of a chemically synthesized miRNA duplex mimic of hsa-microRNA-422b (hsa-miR-422b) (Applied Biosystems) for quantitative normalization of miRNA plasma levels. Total RNA extraction was performed using a microRNA extraction kit (Benevbio), and each sample was eluted in 30 μl H2O. From each plasma extraction, 3 μl (of the 30 μl volume) was used as a template for miRNA reverse transcription (Taqman MicroRNA Reverse Transcription Kit, Applied Biosystems) in a total reaction volume of 15 μl. Following reverse transcription, 2 µl (of the 15 μl volume) of the RT product were then used as template in QPCR with a total reaction volume of 20 µl.
RNA extraction from organ tissue of mice
Mouse organs were harvested and flash frozen in liquid N2. Specifically for stomach preparations, stomach contents were removed and stomach tissue was washed with sterile PBS ten times prior to flash freezing. Frozen tissues were homogenized in Qiazol (Qiagen) using an ultrasonic probe as previously described.Citation30 RNA was then extracted as previously describedCitation64 by the miRNeasy kit (Qiagen) using phenol-chloroform extraction, and column-based extraction. Total RNA concentration was calculated by Nanodrop spectrophotometry (ThermoScientific). To optimize the ability to detect low levels of miRNAs, a relatively large amount of total RNA (60 ng of total RNA) was used for miRNA quantification.
Honey bee tissue collection
Nurses and foragers were collected from the same bee colonies at identical times using a mouth aspirator. Nurses were collected from the brood area of frames and were identified as nurses after they repeatedly placed their heads into honeycomb cells containing larvae. Foragers were collected from the front of the hive or on the landing board as they returned to the hive with visible loads of pollen on their legs.Citation65 Gut tissue was removed from abdomens, and the remaining abdominal tissue was also independently used for gene expression analysis. All dissected material was placed into RNAlater (Invitrogen) for storage prior to analysis. RNA was then extracted as previously describedCitation64 by the miRNeasy kit (Qiagen) using phenol-chloroform extraction, and column-based extraction. Total RNA concentration was calculated by Nanodrop spectrophotometry (ThermoScientific). To optimize the ability to detect low levels of miRNAs, a relatively large amount of total RNA (300 ng of total RNA) was used for miRNA quantification.
RNA extraction from food
Ripened avocado, apple, banana, orange and cantaloupe as well as processed ham were purchased from a commercial retailer and were marketed for human consumption. For fruit products, rind and skin were discarded and fruit flesh was processed for analysis. A standard mass of food (1.5 g) was added to Qiazol (1:9, food weight in g/Qiazol volume in ml). For pollen, 100 mg was added to Qiazol (1:9, 100 mg pollen/900 μl Qiazol). For honey, 100 μl was added to Qiazol (1:9, 100 ul honey/900 μl Qiazol). To that mixture, 2 ficomoles were added of a chemically synthesized miRNA duplex mimic of hsa-microRNA-422b (hsa-miR-422b) (Ambion/Applied Biosystems) for quantitative normalization of miRNA plasma levels. Food was then homogenized using mechanical disruption. RNA was then extracted as previously describedCitation64 by the miRNeasy kit (Qiagen) using phenol-chloroform extraction and column-based extraction. As with plasma, each pollen and honey sample was eluted in 30 μl H2O. For all other food products, each sample was eluted in 30 μl H2O; subsequently, a 1:10 dilution was made into 30 μl H2O for use in reverse transcription. Specifically, 3 μl (of the 30 μl volume) was used as a template for miRNA reverse transcription (Taqman MicroRNA Reverse Transcription Kit, Applied Biosystems) in a total reaction volume of 15 μl. Following reverse transcription, 2 µl (of the 15 μl volume) of the RT product was then used as template in QPCR with a total reaction volume of 20 µl.
Quantification of microRNA and determination of copy number
To quantify levels of select miRNAs, standard Taqman reverse transcription-quantitative (“real-time”) polymerase chain reaction (RT-QPCR) was utilized, as we previously described.Citation64 Taqman primer/probes for quantification were used as follows (Applied Biosystems): dme-let7 [AB 4427975 (000332)], hsa-miR-16 [AB 4427975 (000391)], hsa-miR-21 [AB 4427975 (000397)], has-miR-195 [AB 4427975 (002107)], ath-MIR156a [AB 4427975 (000333)], ath-MIR159a [AB 4427975 (000338)], ath-MIR-169a [AB 4427975 (000352)] and hsa-miR-422b [AB 4427975 (000575)]. In doing so, raw Ct values were generated, representing the “real-time” cycle number at which the increase in miRNA probe fluorescence is exponential. As previously described,Citation66 mature miRNA copy numbers were then calculated from raw Ct values through generation of individual standard curves for each miRNA Taqman primer/probe set after making serial dilutions of a known quantity of synthetic oligonucleotide mimic for each mature miRNA (Applied Biosystems). On average, the level of detection for a miRNA by this method was approximately three copies in a given reaction, consistent with prior reports.Citation66 Subsequently, for plasma samples, copy number was calculated per μl of plasma tested. For bee and organ tissue, copy number was calculated per 10 pg of total RNA tested (10 pg of RNA representing a conservative estimate of the total RNA content of a single eukaryotic cell). For stomach contents, copy number was calculated from total stomach content per individual mouse.
To control for potential inconsistencies in miRNA extraction from plasma, food or stomach contents when analyzing extracellular or ex vivo miRNAs, copy numbers of miRNAs in plasma, food or stomach contents were normalized (e.g., “normalized copy number”) using a ratio calculated from levels of a miR-422b reference control exogenously spiked into each given sample prior to extraction as compared with an equivalent amount of miR-422b spiked in water (“gold standard”) prior to extraction. Furthermore, in plasma samples, endogenous levels of miR-16 were also quantified as a positive control (Fig. S1). Notably, in food and plasma samples as previously described,Citation31 endogenous miR-422b is expressed at substantially lower or absent levels as compared with the exogenously added controls, thus not confounding this normalization method. Alternatively, as a reference control for miRNA quantification in mouse organ tissue, endogenously expressed miR-195 was assessed to confirm consistent patterns of expression across dietary groups. MiR-195 was empirically chosen, given its abundant expression in many tissues and no known functional relationship to miR-21 or any of the plant miRNAs analyzed in this study (Fig. S1). Notably, there is no known housekeeping miRNA reported in honey bee tissue to our knowledge that is consistently expressed among nurses and foragers. Thus, as reference control for miRNA quantification, the β-actin housekeeping gene was used to confirm consistent patterns of expression across the abdominal tissue of nurses and foragers, as β-actin has been shown not to differ between the nurse and forager stages (60).
Statistical analysis
Unless otherwise indicated, all numerical quantifications represent mean ± standard error of the mean (n, number of independent experimental repetitions or number of independent animals). Paired samples were compared by Student’s t-test. Comparison of multiple samples was performed by one-way ANOVA followed by Student Newman-Keuls post hoc tests to calculate p values. Values of p < 0.05 are considered significant.
| Abbreviations: | ||
| AGO2 | = | Argonaute 2 |
| dsRNA | = | double-stranded RNA |
| miRNA | = | microRNA |
| OJ | = | orange juice |
| oz | = | ounce |
Additional material
Download Zip (161.8 KB)Acknowledgments
We thank Dr S.K. Chan (critical advice and reading of manuscript) and S. Tribuna (administrative assistance). This work was supported by the National Institutes of Health (K08HL096834 to S.Y.C.); the Lerner, Harris, Radovsky-McArthur and Watkins Foundations (S.Y.C.); Gilead Sciences (Gilead Research Scholars Fund to S.Y.C.) and the Pulmonary Hypertension Association (S.Y.C.).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Zhang Y, Liu D, Chen X, Li J, Li L, Bian Z, et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell 2010; 39:133 - 44; http://dx.doi.org/10.1016/j.molcel.2010.06.010; PMID: 20603081
- Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem 2010; 285:17442 - 52; http://dx.doi.org/10.1074/jbc.M110.107821; PMID: 20353945
- Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci USA 2010; 107:6328 - 33; http://dx.doi.org/10.1073/pnas.0914843107; PMID: 20304794
- Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol 2011; 13:423 - 33; http://dx.doi.org/10.1038/ncb2210; PMID: 21423178
- Mittelbrunn M, Gutiérrez-Vázquez C, Villarroya-Beltri C, González S, Sánchez-Cabo F, González MA, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun 2011; 2:282; http://dx.doi.org/10.1038/ncomms1285; PMID: 21505438
- Hergenreider E, Heydt S, Tréguer K, Boettger T, Horrevoets AJ, Zeiher AM, et al. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat Cell Biol 2012; 14:249 - 56; http://dx.doi.org/10.1038/ncb2441; PMID: 22327366
- Zhuang G, Wu X, Jiang Z, Kasman I, Yao J, Guan Y, et al. Tumour-secreted miR-9 promotes endothelial cell migration and angiogenesis by activating the JAK-STAT pathway. EMBO J 2012; 31:3513 - 23; http://dx.doi.org/10.1038/emboj.2012.183; PMID: 22773185
- Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007; 9:654 - 9; http://dx.doi.org/10.1038/ncb1596; PMID: 17486113
- Gupta SK, Bang C, Thum T. Circulating microRNAs as biomarkers and potential paracrine mediators of cardiovascular disease. Circ Cardiovasc Genet 2010; 3:484 - 8; http://dx.doi.org/10.1161/CIRCGENETICS.110.958363; PMID: 20959591
- Jung M, Schaefer A, Steiner I, Kempkensteffen C, Stephan C, Erbersdobler A, et al. Robust microRNA stability in degraded RNA preparations from human tissue and cell samples. Clin Chem 2010; 56:998 - 1006; http://dx.doi.org/10.1373/clinchem.2009.141580; PMID: 20378769
- Asgari S. MicroRNA functions in insects. Insect Biochem Mol Biol 2013; 43:388 - 97; http://dx.doi.org/10.1016/j.ibmb.2012.10.005; PMID: 23103375
- Lucas K, Raikhel AS. Insect microRNAs: biogenesis, expression profiling and biological functions. Insect Biochem Mol Biol 2013; 43:24 - 38; http://dx.doi.org/10.1016/j.ibmb.2012.10.009; PMID: 23165178
- Huvenne H, Smagghe G. Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: a review. J Insect Physiol 2010; 56:227 - 35; http://dx.doi.org/10.1016/j.jinsphys.2009.10.004; PMID: 19837076
- Price DRG, Gatehouse JA. RNAi-mediated crop protection against insects. Trends Biotechnol 2008; 26:393 - 400; http://dx.doi.org/10.1016/j.tibtech.2008.04.004; PMID: 18501983
- Tomoyasu Y, Miller SC, Tomita S, Schoppmeier M, Grossmann D, Bucher G. Exploring systemic RNA interference in insects: a genome-wide survey for RNAi genes in Tribolium. Genome Biol 2008; 9:R10; http://dx.doi.org/10.1186/gb-2008-9-1-r10; PMID: 18201385
- Saleh MC, Tassetto M, van Rij RP, Goic B, Gausson V, Berry B, et al. Antiviral immunity in Drosophila requires systemic RNA interference spread. Nature 2009; 458:346 - 50; http://dx.doi.org/10.1038/nature07712; PMID: 19204732
- Jarosch A, Moritz RFA. Systemic RNA-interference in the honeybee Apis mellifera: tissue dependent uptake of fluorescent siRNA after intra-abdominal application observed by laser-scanning microscopy. J Insect Physiol 2011; 57:851 - 7; http://dx.doi.org/10.1016/j.jinsphys.2011.03.013; PMID: 21439290
- Zhang L, Hou D, Chen X, Li D, Zhu L, Zhang Y, et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res 2012; 22:107 - 26; http://dx.doi.org/10.1038/cr.2011.158; PMID: 21931358
- Zhang Y, Wiggins BE, Lawrence C, Petrick J, Ivashuta S, Heck G. Analysis of plant-derived miRNAs in animal small RNA datasets. BMC Genomics 2012; 13:381; http://dx.doi.org/10.1186/1471-2164-13-381; PMID: 22873950
- Jiang M, Sang X, Hong Z. Beyond nutrients: food-derived microRNAs provide cross-kingdom regulation. Bioessays 2012; 34:280 - 4; http://dx.doi.org/10.1002/bies.201100181; PMID: 22354805
- Chen X, Liang H, Zhang J, Zen K, Zhang CY. Horizontal transfer of microRNAs: molecular mechanisms and clinical applications. Protein Cell 2012; 3:28 - 37; http://dx.doi.org/10.1007/s13238-012-2003-z; PMID: 22314808
- Liang H, Huang L, Cao J, Zen K, Chen X, Zhang CY. Regulation of mammalian gene expression by exogenous microRNAs. Wiley Interdiscip Rev RNA 2012; 3:733 - 42; http://dx.doi.org/10.1002/wrna.1127; PMID: 22740375
- Liang H, Zen K, Zhang J, Zhang CY, Chen X. New roles for microRNAs in cross-species communication. RNA Biol 2013; 10:373 - 6; http://dx.doi.org/10.4161/rna.23663; PMID: 23364352
- Igaz P, Nagy Z, Vásárhelyi B, Buzás E, Falus A, Rácz K. [Potential role for microRNAs in inter-individual and inter-species communication]. Orv Hetil 2012; 153:1647 - 50; http://dx.doi.org/10.1556/OH.2012.29463; PMID: 23063896
- Vaucheret H, Chupeau Y. Ingested plant miRNAs regulate gene expression in animals. Cell Res 2012; 22:3 - 5; http://dx.doi.org/10.1038/cr.2011.164; PMID: 22025251
- Witzany G. Do we eat gene regulators?. Commun Integr Biol 2012; 5:230 - 2; http://dx.doi.org/10.4161/cib.19621; PMID: 22896781
- Baum JA, Bogaert T, Clinton W, Heck GR, Feldmann P, Ilagan O, et al. Control of coleopteran insect pests through RNA interference. Nat Biotechnol 2007; 25:1322 - 6; http://dx.doi.org/10.1038/nbt1359; PMID: 17982443
- Mao YB, Cai WJ, Wang JW, Hong GJ, Tao XY, Wang LJ, et al. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat Biotechnol 2007; 25:1307 - 13; http://dx.doi.org/10.1038/nbt1352; PMID: 17982444
- Xie Z, Allen E, Fahlgren N, Calamar A, Givan SA, Carrington JC. Expression of Arabidopsis MIRNA genes. Plant Physiol 2005; 138:2145 - 54; http://dx.doi.org/10.1104/pp.105.062943; PMID: 16040653
- Parikh VN, Jin RC, Rabello S, Gulbahce N, White K, Hale A, et al. MicroRNA-21 integrates pathogenic signaling to control pulmonary hypertension: results of a network bioinformatics approach. Circulation 2012; 125:1520 - 32; http://dx.doi.org/10.1161/CIRCULATIONAHA.111.060269; PMID: 22371328
- Baggish AL, Hale A, Weiner RB, Lewis GD, Systrom D, Wang F, et al. Dynamic regulation of circulating microRNA during acute exhaustive exercise and sustained aerobic exercise training. J Physiol 2011; 589:3983 - 94; http://dx.doi.org/10.1113/jphysiol.2011.213363; PMID: 21690193
- Wang C, Han J, Liu C, Kibet KN, Kayesh E, Shangguan L, et al. Identification of microRNAs from Amur grape (Vitis amurensis Rupr.) by deep sequencing and analysis of microRNA variations with bioinformatics. BMC Genomics 2012; 13:122; http://dx.doi.org/10.1186/1471-2164-13-122; PMID: 22455456
- Grant-Downton R, Le Trionnaire G, Schmid R, Rodriguez-Enriquez J, Hafidh S, Mehdi S, et al. MicroRNA and tasiRNA diversity in mature pollen of Arabidopsis thaliana. BMC Genomics 2009; 10:643; http://dx.doi.org/10.1186/1471-2164-10-643; PMID: 20042113
- Kirschner MB, Kao SC, Edelman JJ, Armstrong NJ, Vallely MP, van Zandwijk N, et al. Haemolysis during sample preparation alters microRNA content of plasma. PLoS One 2011; 6:e24145; http://dx.doi.org/10.1371/journal.pone.0024145; PMID: 21909417
- Sambrook J, Russell DW. Molecular Cloning: a Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press, 2001.
- Brown BD, Gentner B, Cantore A, Colleoni S, Amendola M, Zingale A, et al. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat Biotechnol 2007; 25:1457 - 67; http://dx.doi.org/10.1038/nbt1372; PMID: 18026085
- Wilson DS, Dalmasso G, Wang L, Sitaraman SV, Merlin D, Murthy N. Orally delivered thioketal nanoparticles loaded with TNF-α-siRNA target inflammation and inhibit gene expression in the intestines. Nat Mater 2010; 9:923 - 8; http://dx.doi.org/10.1038/nmat2859; PMID: 20935658
- Lynch M. The Origins of Genome Architecture. Sunderland: Sinauer Associates, Inc. Publishers, 2007.
- Moxon S, Jing R, Szittya G, Schwach F, Rusholme Pilcher RL, Moulton V, et al. Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res 2008; 18:1602 - 9; http://dx.doi.org/10.1101/gr.080127.108; PMID: 18653800
- Zuo J, Zhu B, Fu D, Zhu Y, Ma Y, Chi L, et al. Sculpting the maturation, softening and ethylene pathway: the influences of microRNAs on tomato fruits. BMC Genomics 2012; 13:7; http://dx.doi.org/10.1186/1471-2164-13-7; PMID: 22230737
- Boeing H, Bechthold A, Bub A, Ellinger S, Haller D, Kroke A, et al. Critical review: vegetables and fruit in the prevention of chronic diseases. Eur J Nutr 2012; 51:637 - 63; http://dx.doi.org/10.1007/s00394-012-0380-y; PMID: 22684631
- Izumi H, Kosaka N, Shimizu T, Sekine K, Ochiya T, Takase M. Bovine milk contains microRNA and messenger RNA that are stable under degradative conditions. J Dairy Sci 2012; 95:4831 - 41; http://dx.doi.org/10.3168/jds.2012-5489; PMID: 22916887
- Munch EM, Harris RA, Mohammad M, Benham AL, Pejerrey SM, Showalter L, et al. Transcriptome profiling of microRNA by Next-Gen deep sequencing reveals known and novel miRNA species in the lipid fraction of human breast milk. PLoS One 2013; 8:e50564; http://dx.doi.org/10.1371/journal.pone.0050564; PMID: 23418415
- Admyre C, Johansson SM, Qazi KR, Filén JJ, Lahesmaa R, Norman M, et al. Exosomes with immune modulatory features are present in human breast milk. J Immunol 2007; 179:1969 - 78; PMID: 17641064
- Kosaka N, Izumi H, Sekine K, Ochiya T. microRNA as a new immune-regulatory agent in breast milk. Silence 2010; 1:7; http://dx.doi.org/10.1186/1758-907X-1-7; PMID: 20226005
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 2001; 263:103 - 12; http://dx.doi.org/10.1016/S0378-1119(00)00579-5; PMID: 11223248
- Yu N, Christiaens O, Liu J, Niu J, Cappelle K, Caccia S, et al. Delivery of dsRNA for RNAi in insects: an overview and future directions. Insect Sci 2012; 20:4 - 14; http://dx.doi.org/10.1111/j.1744-7917.2012.01534.x
- Amdam GV, Simões ZL, Guidugli KR, Norberg K, Omholt SW. Disruption of vitellogenin gene function in adult honeybees by intra-abdominal injection of double-stranded RNA. BMC Biotechnol 2003; 3:1; http://dx.doi.org/10.1186/1472-6750-3-1; PMID: 12546706
- Aronstein K, Pankiwi T, Saldivar E. SID-I is implicated in systemic gene silencing in the honey bee. J Apic Res 2006; 45:20 - 4
- Maori E, Paldi N, Shafir S, Kalev H, Tsur E, Glick E, et al. IAPV, a bee-affecting virus associated with Colony Collapse Disorder can be silenced by dsRNA ingestion. Insect Mol Biol 2009; 18:55 - 60; http://dx.doi.org/10.1111/j.1365-2583.2009.00847.x; PMID: 19196347
- Nunes FM, Simões ZL. A non-invasive method for silencing gene transcription in honeybees maintained under natural conditions. Insect Biochem Mol Biol 2009; 39:157 - 60; http://dx.doi.org/10.1016/j.ibmb.2008.10.011; PMID: 19049870
- Hunter W, Ellis J, Vanengelsdorp D, Hayes J, Westervelt D, Glick E, et al. Large-scale field application of RNAi technology reducing Israeli acute paralysis virus disease in honey bees (Apis mellifera, Hymenoptera: Apidae). PLoS Pathog 2010; 6:e1001160; http://dx.doi.org/10.1371/journal.ppat.1001160; PMID: 21203478
- Liu X, Zhang Y, Yan X, Han R. Prevention of Chinese sacbrood virus infection in Apis cerana using RNA interference. Curr Microbiol 2010; 61:422 - 8; http://dx.doi.org/10.1007/s00284-010-9633-2; PMID: 20379718
- Desai SD, Eu YJ, Whyard S, Currie RW. Reduction in deformed wing virus infection in larval and adult honey bees (Apis mellifera L.) by double-stranded RNA ingestion. Insect Mol Biol 2012; 21:446 - 55; http://dx.doi.org/10.1111/j.1365-2583.2012.01150.x; PMID: 22690671
- Nazzi F, Brown SP, Annoscia D, Del Piccolo F, Di Prisco G, Varricchio P, et al. Synergistic parasite-pathogen interactions mediated by host immunity can drive the collapse of honeybee colonies. PLoS Pathog 2012; 8:e1002735; http://dx.doi.org/10.1371/journal.ppat.1002735; PMID: 22719246
- Garbian Y, Maori E, Kalev H, Shafir S, Sela I. Bidirectional transfer of RNAi between honey bee and Varroa destructor: Varroa gene silencing reduces Varroa population. PLoS Pathog 2012; 8:e1003035; http://dx.doi.org/10.1371/journal.ppat.1003035; PMID: 23308063
- Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet 2011; 12:19 - 31; http://dx.doi.org/10.1038/nrg2916; PMID: 21116305
- Crailsheim K, Schneider L, Hrassnigg N, Buhlmann G, Brosch U, Gmeinbauer R, et al. Pollen consumption and utilization in worker honeybees (Apis mellifera carnica): dependence on individual age and function. J Insect Physiol 1992; 38:409 - 19; http://dx.doi.org/10.1016/0022-1910(92)90117-V
- Rortais A, Arnold G, Halm M-P, Touffet-Briens F. Modes of honeybees exposure to systemic insecticides: estimated amounts of contaminated pollen and nectar consumed by different categories of bees. Apidologie (Celle) 2005; 36:71 - 83; http://dx.doi.org/10.1051/apido:2004071
- Shi YY, Wu XB, Huang ZY, Wang ZL, Yan WY, Zeng ZJ. Epigenetic modification of gene expression in honey bees by heterospecific gland secretions. PLoS One 2012; 7:e43727; http://dx.doi.org/10.1371/journal.pone.0043727; PMID: 22928024
- Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA 2012; 109:E2110 - 6; http://dx.doi.org/10.1073/pnas.1209414109; PMID: 22753494
- van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med 2013; 368:407 - 15; http://dx.doi.org/10.1056/NEJMoa1205037; PMID: 23323867
- Lu TX, Hartner J, Lim EJ, Fabry V, Mingler MK, Cole ET, et al. MicroRNA-21 limits in vivo immune response-mediated activation of the IL-12/IFN-gamma pathway, Th1 polarization, and the severity of delayed-type hypersensitivity. J Immunol 2011; 187:3362 - 73; http://dx.doi.org/10.4049/jimmunol.1101235; PMID: 21849676
- Chan SY, Zhang YY, Hemann C, Mahoney CE, Zweier JL, Loscalzo J. MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metab 2009; 10:273 - 84; http://dx.doi.org/10.1016/j.cmet.2009.08.015; PMID: 19808020
- Ben-Shahar Y, Robichon A, Sokolowski MB, Robinson GE. Influence of gene action across different time scales on behavior. Science 2002; 296:741 - 4; http://dx.doi.org/10.1126/science.1069911; PMID: 11976457
- Schmittgen TD, Lee EJ, Jiang J, Sarkar A, Yang L, Elton TS, et al. Real-time PCR quantification of precursor and mature microRNA. Methods 2008; 44:31 - 8; http://dx.doi.org/10.1016/j.ymeth.2007.09.006; PMID: 18158130