Abstract
Long non-coding RNAs (lncRNAs) are transcripts longer than ~200 nucleotides with little or no protein-coding capacity. Growing evidence shows that lncRNAs present important function in development and are associated with many human diseases such as cancers, Alzheimer disease, and heart diseases. Transcribed ultraconserved region (T-UCR) transcripts are a novel class of lncRNAs transcribed from ultraconserved regions (UCRs). UCRs are absolutely conserved (100%) between the orthologous regions of the human, rat, and mouse genomes. The UCRs are frequently located at fragile sites and at genomic regions involved in cancers. Recent data suggest that T-UCRs are altered at the transcriptional level in human tumorigenesis and the aberrant T-UCRs expression profiles can be used to differentiate human cancer types. The profound understanding of T-UCRs can throw new light on the pathogenesis of human cancers.
Introduction
The production of RNAs from genomic DNAs is directed by sequences that determine the start and end of transcripts and splicing into mature RNAs.Citation1 Although 93% of human genome can be transcribed into RNAs, only 2% of these products can be translated into proteins. The rest of the RNAs show low capability or no capability of being translated into protein, termed as non-coding RNAs (ncRNAs), including rRNAs, microRNAs, tRNAs, mitochondrial ncRNAs,Citation2 small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs), interference RNAs (RNAi), and many long intergenic ncRNAs (lincRNAs).Citation3-Citation5 They can be divided into short non-coding RNAs and long non-coding RNAs (lncRNAs) according to their transcript sizes. The lncRNAs are considered to be longer than 200 nt, making up about 80% of the mammalian ncRNAs.1 Scientists used to ignore the importance of ncRNAs and ncRNAs were classified as “dark matter.” But with the profound research and deep understanding, the significance of ncRNAs has been increasingly emphasized.
The most widely studied class of ncRNAs are miRNAs, which are small ncRNAs of ~22 nucleotides but regulate 60% of coding genes by binding to mRNAs, and are involved in proliferation, differentiation, apoptosis, and development.Citation6 More than 1000 miRNAs have been identified. Their functions and mechanisms in regulation are being continuously discovered.
lncRNAs, as a transcriptional subclass, were first described during the large-scale sequencing of full-length cDNA libraries in the mouse. Many identified lncRNAs were transcribed by RNA polymerase II. Given their unexpected abundance, lncRNAs were initially thought to be spurious “transcriptional noise.” But recent research shows that they take part in various important regulation processes in human and other species. One way to find order in the plethora of reported lncRNAs is to classify them according to genomic location. Thus, lncRNAs can be divided into five categories, namely, antisense lncRNAs, intonic transcripts, lincRNAs, promoter-associated lncRNAs, and UTR-associated lncRNAs (T-UCRs).Citation7 lncRNAs can act as molecular guides, directing chromatin-modifying complexes into specific genomic loci to exert repressive or activating effect on gene expression. They can also transcriptionally or post-transcriptionally regulate gene expression by diverse molecular mechanisms.Citation8 Due to these various functions, lncRNAs play key regulatory roles in physiological processes, especially in cancer biology. lncRNAs are dysregulated in different kinds of cancers, and the expression levels of certain lncRNAs are associated with recurrence, metastasis, and prognosis of cancer. It has been shown that overexpression of certain lncRNAs, behaving like oncogenes, can promote matrix invasion of cancer cells and tumor growth.Citation9
T-UCR transcripts are a novel class of lncRNAs transcribed from ultraconserved region. UCRs are 481 segments that are longer than 200 bp and are 100% conserved in human, rat, and mouse genome, most of which are also identified in chickens and dogs.Citation10 Their transcriptional products, called T-UCRs, constitute a new category of lncRNAs. The T-UCRs expression levels show both a ubiquitous and a tissue-specific pattern. Most importantly, recent data suggests that T-UCRs are altered at the transcriptional level in human tumorigenesis and the aberrant T-UCRs expression profiles can be used to differentiate human cancer types.Citation11
Thus, this article aims to review the current scientific research about T-UCRs in human diseases. Improved understanding of T-UCRs in the diseases could lead to novel prevention strategies, early detection, and improved therapeutics to the diseases.
Ultraconserved Region
UCRs include 481 conserved sequences longer than 200 bp that are absolutely conserved (100%) between orthologous regions of the human, rat, and mouse genomes. These 481 ultraconserved elements were classified into three types, namely, exonic, non-exonic, and possibly exonic.Citation10 Recently, Mestdagh et al.Citation12 re-annotated all UCRs sequences using the more recent genome built hg18. They reorganized UCRs into five different categories, namely, intergenic, intronic, exonic, partly exonic, and exon-containing. () For a few UCRs, the genomic annotation varied because of host gene splice variants. These UCRs were categorized as “multiple.”
Figure 1. Re-annotation of UCRs according to their genomic location with respect to protein-coding genes defined by Refseq.
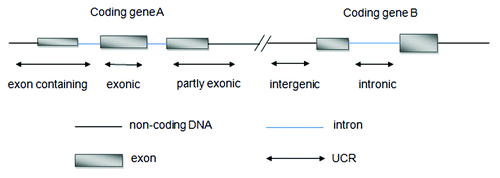
Different tissues have specific UCRs signatures and UCRs are frequently located at fragile sites and cancer-associated genomic regions (CAGRs). In addition to their extreme conservation for the last 400 million years, it has been postulated that these regions could be candidate genes for cancer susceptibility.Citation11 A majority of UCRs can be transcribed, which are considered as T-UCRs. Therefore, it is possible to differentiate human cancers by studying the T-UCRs aberrant expression profiles.
Regulation Mechanisms of T-UCR
So far, the aberrant regulation of T-UCRs expression in cancer has been found to occur in two main ways: by altered interactions with miRNAs and by hypermethylation of CpG island promoters.Citation13 Some T-UCRs have significant antisense complementarity with miRNAs, which could lead to the formation of T-UCR:miRNA pairs. These pairs could impair T-UCRs function by serving a sponge that traps T-UCRs.Citation11 Epigenetic mechanisms can also regulate the expression of T-UCRs. Some T-UCRs are associated with CpG islands in the promoter region of their own host genes.Citation14 The hypermethylation of CpG islands could inhibit the expression of T-UCRs, much like silencing the expression of miRNAs.Citation15
Mestdagh et al.Citation12 gained further insight into the regulation of T-UCRs transcription. They found the transcription of T-UCRs to be located within protein-coding genes was associated with histone marks for active transcription.
T-UCR: miRNA Interaction
uc.160, uc.346A, and uc.348 have significant antisense complementarity with seven miRNAs. Based on the rule of 5′-end “6 base seed,” these T-UCRs could combine with miRNAs. Furthermore, the expression of these T-UCRs was negatively correlated with the miRNAs expression. It can be inferred that these miRNA-T-UCR pairs could inhibit the expression of T-UCRs, in the way similar to miRNA–mRNA interactions. () This postulation was corroborated both in vitro and in vivo. In vitro, uc.160, uc.346A, and uc.348 were cloned into luciferase reporter vectors to assess their interactions with miR-155, miR-24-1, and miR-29-b. Reduction in luciferase expression with four miRNA-T-UCR pairs was observed. In vivo, miR-155 was transfected into MEG01 leukemia cells and the expression levels of uc.346A and uc.160 were significantly reduced after 48 h. Therefore, T-UCRs represented possible targets of miRNAs.Citation11
Table 1. Illustrative list of negative association between T-UCRs and complementary miRNAs in CLLCitation11
In neuroblastoma, Scaruffi et al.Citation16 also demonstrated a negative regulation of T-UCRs by direct interactions with miRNAs. For example, the expression levels of five miRNAs were found to be negatively correlated with expression of nine complementary T-UCRs. These results were in agreement with complementary regulatory mechanisms between miRNAs and T-UCRs ().
Table 2. Illustrative list of negative association between T-UCRs and their complementary miRNAs in neuroblastomaCitation16
T-UCR and Epigenetic Modification
As we know, the downregulation of protein-coding genes, such as tumor suppressor genes in human cancersCitation17 and miRNAs with growth-inhibitory functions,Citation18 have been tightly linked to the presence of CpG island promoter hypermethylation. It is likely that T-UCRs also undergo epigenetic modification.
Three T-UCRs (uc.283A, uc.346, uc.160) had one CpG island within a 2000-bp distance upstream. In HCT-116 (human colorectal cancer cell line), dense methylation was found in their CpG islands; however, these CpG islands were always unmethylated in normal tissues. After the treatment with DNA-demethylating agent, uc.283A, uc.346, and uc.160 restored their expression levels in HCT-116 cell line, which were originally not detectable. It was also confirmed in HCT116 DKO, an isogenic HCT-116 cell line. Moreover, uc.160, uc.283A, and uc.346 CpG islands demethylation featured more accessible chromatin conformation to the enzyme of RNA polymerase II, were also occupied by RNA polymerase II, and accompanied by the presence of trimethylation of lysine 4 of histone H3. The presence of uc.160, uc.283A, and uc.346 CpG island hypermethylation and transcriptional silencing also existed in other cell lines, such as breast (MCF-7, MDA-MB-231, CAMA-1), lung (H552, H441, H358, EBC1), lymphoma (Ramos, Raji, Namalwa), and leukemia cells (HL-60, Jurkat, KG-1a), which reinforced the link between CpG island hypermethylation and T-UCRs silencing.Citation14
Hudson et al.Citation19 studied LNCaP cells treated with 5-AzaC (a DNA hypomethylating agent) and/or TSA (a histone deacetylase inhibitor) for 36 h. They identified six T-UCRs, uc.308A, uc.434A, uc.241A, uc.283A, uc.285, and uc.85, were consistently upregulated in the three treatment groups (5-AzaC only, TSA only, and the combination). Their results also indicated that these T-UCRs were epigenetically silenced in prostate cancer. Among them was uc.283A, which was previously found to be silenced by CpG hypermethylation in human colon cancer cells.
T-UCR and Histone Marks
Mestdagh et al.Citation12 evaluated the H3K4me3 distance distributions of intergenic and intragenic T-UCRs. Both intergenic and intragenic T-UCRs were significantly associated with active trimethylation of lysine 4 of histone H3 (H3K4me3), a marker for transcriptional initiation, but with a different distribution as compared with protein-coding genes, suggesting a difference in transcriptional organization between T-UCRs and protein-coding genes. In addition, miRNAs were closely associated with active H3K4me3 marks. The genomic location of H3K4me3 marks for miRNAs and T-UCRs appeared similar, suggesting common features of transcription organization for these two classes of ncRNAs.
T-UCR in the Disease
Recently, various detective techniques and genome-wide microarray profiling have showed that UCRs are frequently transcribed and there are distinct signatures in human carcinomas,Citation11 including hepatocellular carcinoma,Citation20 prostate cancer,Citation19 neuroblastoma,Citation12 colorectal carcinoma,Citation11,Citation21 and chronic lymphocytic leukemia.Citation11 Calin et al.Citation11 were the first to investigate the expression of T-UCRs in human cancers. They found the malignant cells had a unique spectrum of T-UCRs when compared with normal cells, which suggested that significant variations in T-UCRs expression were involved in the malignant process. The altered transcriptional levels in human tumorigenesis and the aberrant T-UCRs expression profiles indicate novel mechanisms in the progress of multiple cancers.
T-UCR in Hepatocellular Carcinoma
There were 56 identified T-UCRs that were aberrantly and significantly expressed in malignant HepG2 cells compared with non-malignant human hepatocytes, with 33 T-UCRs increased and 23 decreased. The most significant change was noted for uc.338, which was increased in expression by nearly 7-fold. Besides, there was a significant difference in uc.338 expression between tumoral and adjacent non-tumoral tissues.Citation20
To gain insight into the functional role of uc.338, Braconi et al.Citation20 performed gene annotation enrichment analysis of genome-wide mRNAs that were changed in expression after uc.338 inhibition by siRNA. The results indicated predominant effects on genes involved in cell cycle progression from phase G1 to phase S. These effects were experimentally demonstrated in both human and murine cells. The inhibition of uc.338 decreased both anchorage-dependent and anchorage-independent growth of HCC cells. Moreover, inhibition of uc.338 in HCC reduced the number of cells in S phase, because the inhibition of uc.338 induced an increased expression of tumor suppressor p16INK4a and the reduction of CDK4, CDK6, and cyclin D1, which participated in the modulation of S-phase progression.
Interestingly, all cholangiocarcinoma cells showed quite low levels of uc.338 expression, suggesting that uc.338 may differentiate between primary liver carcinomas arising from different liver epithelia.Citation20 Thus, uc.338 was increased in HCC cells and might be a promising marker for HCC.
T-UCR in the Prostate Cancer
Hudson et al.Citation19 also showed T-UCRs were aberrantly expressed in prostate cancer. Systemic analysis of microarrays showed that numerous T-UCRs were found to be differentially expressed between tumor and non-cancerous tissue between tumors with high-Gleason grade (≥7) and low-Gleason grade (≤6), and between tumors with metastasis and those without. Exploratory prediction analysis of microarrays identified 60 T-UCRs, which could robustly separate non-tumor tissues from tumor tissues. uc.363A and uc.454A were the most significantly upregulated and downregulated T-UCRs, respectively.
Only few ucRNAs were differently expressed in different stages of the prostate tumors. For example, uc.106A was upregulated in prostate tumors, but tended to be downregulated in tumors with high-Gleason grade and extraprostatic extension. siRNA-targeting uc.106A was transfected into LNCaP cells to probe into its functional role. Global expression analysis found numerous genes were upregulated and downregulated in the siRNA-transfected cells. These genes were clustered in distinct pathways related to proliferation, cell movement, cancer, and immune response. It indicated that uc.106 may affect cellular transcription pattern in prostate cancer cells.Citation19
T-UCR in the Neuroblastoma
Neuroblastoma patients present with a highly variable clinical course and MYCN -amplification usually indicates an aggressive tumor with rapid progression and a poor outcome. T-UCRs expression was related to prognosis and MYCN—amplification in neuroblastoma. Seven T-UCRs (uc.347, uc.350, uc.279, uc.460, uc.379, uc.446, and uc.364) were upregulated in MYCN-amplified tumors and no T-UCRs were downregulated. By using the SHEP-MYCN-ER cellular model system, which allowed MYCN activation upon addition of 4-hydroxy tamoxifen, they found that three out of seven T-UCRs (uc.460, uc.350, and uc.379) were increased significantly than the rest four, suggesting they were MYCN-responsive.Citation12
Having established differential T-UCR expression patterns in neuroblastoma tumors, Mestdagh et al.Citation12 also sought to assign putative functions to each T-UCR. By using gene set enrichment analysis (GSEA),Citation22 classes of T-UCRs were found to be widely associated with numerous cancer-related cellular functions and pathways such as proliferation, apoptosis, and differentiation. They sought independent experimental validation for a subset of inferred T-UCR functions. They found that 40 T-UCRs featured TP53-dependent expression. Among 40 altered T-UCRs, almost three-quarters (29 of 40) were annotated to the TP53 response pathway.
Besides the individual aberrant expression of T-UCR, Mestdagh et al.Citation12 also discovered a network of functional T-UCRs in neuroblastoma. T-UCRs that were located in or around genes may share functions. Then four groups of T-UCRs were identified, consisting of nine, 11, nine, and six T-UCRs, respectively (). As noted in GSEA annotation, cluster 1 was linked to DNA damage. Cluster 2 was predominantly associated with cell cycle regulation and proliferation. Cluster 3 appeared to be implicated in differentiation, and cluster 4 was associated with development and immune response. Experimental models were used to validate the annotations of these clusters and the results proved the putative functions of these four clusters.Citation12
Table 3. T-UCR expression network in neuroblastomaCitation12
T-UCR in the Colorectal Cancer
T-UCRs expression profiles were also identified in the colorectal cancer (CRC). uc.73 was one of the frequently upregulated T-UCRs in colon cancer. It possibly had a correlation with the pathogenesis of the CRC. Calin et al.Citation11 investigated the effects of uc.73 downregulation in COLO-320 colorectal cancer cells. SW620 colon cancer cells were used as a control in which the expression of uc.73 does not differ from normal colonic cells. siRNA1, siRNA3, and a pool of four siRNAs were transfected into COLO-320 and SW620 cells to target uc.73. There was significantly less expression of uc.73 in both COLO-320 and SW620 cells after the treatment with siRNAs 1, 3, and pool. Besides, the growth of COLO-320 cells was significantly reduced after the treatment, but not in SW620 cells. Cell cycle studies revealed an increase in the sub-G1 fraction of cells, which suggested the presence of apoptosis in COLO-320 cells. This finding was also confirmed by the apoptosis-specific AnnexinV assay and caspase-3 assay. Furthermore, there was a positive relation between the intensity of effects on cell apoptosis and the degree of inhibition to uc.73. These data suggested that uc.73 could increase the number of malignant cells by reducing apoptosis. Interestingly, Mestdagh et al.Citation12 also found uc.73 annotated to the TP53 response pathway, supporting its reported role in apoptosis.
However, there was also a contradictory report with regards to these findings. Sana et al.Citation21 found significantly lower expression levels of uc.73 and uc.388 in CRC samples compared with control samples. Besides, higher levels of uc.73 expression was correlated with overall survival in CRC patients. No significant associations of uc.73 and uc.388 with clinical stage, grade, and tumor diameter were discovered. These results showed an opposite trend in comparison to the previous study. The reasons may be either that in the first study hybridization microarrays were used, while the second employed real-time PCR, or due to the small number of patients examined in the second study.
T-UCR in the Leukemia
T-UCRs also appeared to be involved in the hematological malignancies. T-UCRs differentially expressed in human cancers were located in cancer-associated genomic regions (CAGRs).Citation11 For example, no mutations have been detected in the chromosomal region of 13q21.33-q22.2 (linked to susceptibility to familial CLL74), but two T-UCRs (uc.349A and uc.352) were encoded within this region. uc.349A was downregulated and uc.352 was upregulated in malignant B-CLL CD5-positive cells. It has been hypothesized that these T-UCRs could be causatively linked to familial disease inheritance.
CLL signature was composed of 19 T-UCRs (eight upregulated and 11 downregulated). Among them, the expression of uc.291 and uc.73A were significantly higher in normal CD5+/CD19+ lymphocytes than in CLL cells. This result was also confirmed in other studies.Citation23-Citation25 Interestingly, uc.73 was demonstrated as an oncogene in colorectal cancer cell lines. The opposite deregulation observed in two different types of cancer suggested a possible tumor-specific function for this T-UCR.Citation26
Therapies Targeting T-UCR
There is increasing interest in the potential involvement of T-UCRs in disease etiology, owing to aberrant function of T-UCRs in differentiation and developmental processes. Expectations in the therapeutic area have been raised as a result of the recognition of T-UCRs defects in human diseases. The ability to regulate aberrant expressed T-UCRs may open new avenues to treat the diseases. Recently, there is little research focusing on T-UCR therapy. The following reflects the hypothesis about the treatment toward T-UCRs. The targeting of T-UCRs is still in its fancy. But with deeper research and understanding of T-UCRs, some effective and innovative regimens will appear in the future.
The upregulation of several T-UCRs was found to be with the risk of occurrence of tumors and the metastases. Therefore, it is possible to use antisense oligonucleotides (ASOs) to inhibit T-UCR functions. The similar method has been used in miRNA inhibition. In a mouse mammary tumor model, the intravenous injection of ASO (targeting miR-10b) prevented the metastasis and suppresses dissemination to the lungs.Citation27 miR-122 was involved in the regulation of cholesterol and lipid metabolism. An ASO that targeted miR-122 induced the inhibition of its function and decreased plasma cholesterol level.Citation28 lncRNA BACE1-AS was identified as upregulated in Alzheimer disease.Citation29 siRNA oligos targeting this lncRNA is under research. Companies and institutions such as the Allen Institute for Brain Science, CuRNA, Regulus Therapeutics, Miragen Therapeutics, and Santaris Pharma are developing ncRNA-based strategies against cancer, cardiovascular, neurological, and muscular diseases.Citation30
Another innovative strategy may inhibit the function of T-UCRs. It has been used to target miRNAs. “miRNA sponges” were vectors containing multiple artificial miRNA binding sites. They produced large quantities of transcript, acting as sponges for cognate miRNAs, preventing their association with natural targets. For example, miRNA sponge was used to inhibit miR-9 in highly malignant cells.Citation31 This method may be useful for the inhibition of T-UCRs, but its feasibility needs to be proven.
The administration of DNA-demethylating agents and histone deacetylase inhibitors could be used to restore the expression of tumor suppressor T-UCRs, thereby stopping tumoral growth and ultimately resulting in the programmed cell death of the transformed cells.Citation14 These agents even without target specificity have shown themselves to have therapeutic benefits and have received clinical approval for the treatment of certain hematological malignancies.Citation32
An adenoviral vector was used to deliver miR-26a, a tumor suppressor miRNA, into a mouse model of hepatocellular carcinoma, resulting in suppression of proliferation and induction of apoptosis. This was called “miRNA replacement therapy.”Citation33 In the future, we can also try to deliver T-UCRs with tumor suppression properties, in order to restore their expression.
Discussion
Four hundred and eighty-one UCR sequences show 100% conservation among the three species, human, rat, and mouse genomes. Some of them contain protein-coding sequences, while over half are not predicted to for any proteins. A subclass of ncRNAs encoded from a subset of UCRs was identified: transcribed UCRs (T-UCRs). Scientists started to investigate the role of T-UCRs in the pathogenesis of multiple diseases. Recent findings strongly support the involvement of T-UCRs in the pathogenesis of cancer.Citation11,Citation12,Citation19-Citation21 So far, the aberrant regulation of T-UCRs expression in cancer has been found to occur in two main ways: by altered interactions with miRNAs and by hypermethylation of CpG island promoters.Citation13 Furthermore, transcription of T-UCRs located within protein-coding genes was associated with histone marks for active transcription.Citation12
T-UCRs are altered at the transcriptional level in human tumorigenesis and the aberrant T-UCRs expression profiles can be used to differentiate cancer behaviors. Calin and his team were the first group to investigate the expression of T-UCRs in human cancers.Citation11 They found that all the malignant cells have unique spectra of expressed T-UCRs compared with the corresponding normal cells, which suggested that variations in T-UCR expression were involved in the malignant process. T-UCRs might offer a novel strategy for cancer diagnosis and prognosis.
A large fraction of T-UCRs are still uncharacterized and the development of strategies to target T-UCRs is still unexplored. But the increasing scientific reports about T-UCRs will offer a promising future. T-UCRs represent attractive therapeutic targets and recent experiments targeting other ncRNAs provide possible ideas for T-UCRs therapies. New developments are expected in this area. Exciting times lie ahead of us.
| Abbreviations: | ||
| T-UCR | = | transcribed ultraconserved region |
| UCR | = | ultraconserved regions |
| ncRNA | = | non-coding RNA |
| lincRNAs | = | long intergenic ncRNAs |
| snoRNAs | = | small nucleolar RNAs |
| RNAi | = | interference RNAs |
| CAGRs | = | ancer-associated genomic regions |
| GSEA | = | gene set enrichment analysis |
| CRC | = | colorectal cancer |
| ASO | = | antisense oligonucleotides |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Financial Disclosure Statement
This work was supported by the National Natural Science Foundation of China (No. 81000161 and No.81170362).
References
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al, FANTOM Consortium, RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group). The transcriptional landscape of the mammalian genome. Science 2005; 309:1559 - 63; http://dx.doi.org/10.1126/science.1112014; PMID: 16141072
- Dermitzakis ET, Reymond A, Antonarakis SE. Conserved non-genic sequences - an unexpected feature of mammalian genomes. Nat Rev Genet 2005; 6:151 - 7; http://dx.doi.org/10.1038/nrg1527; PMID: 15716910
- Costa FF. Non-coding RNAs: lost in translation?. Gene 2007; 386:1 - 10; http://dx.doi.org/10.1016/j.gene.2006.09.028; PMID: 17113247
- Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science 2008; 319:1787 - 9; http://dx.doi.org/10.1126/science.1155472; PMID: 18369136
- Collins LJ, Penny D. The RNA infrastructure: dark matter of the eukaryotic cell?. Trends Genet 2009; 25:120 - 8; http://dx.doi.org/10.1016/j.tig.2008.12.003; PMID: 19171405
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009; 136:215 - 33; http://dx.doi.org/10.1016/j.cell.2009.01.002; PMID: 19167326
- Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics 2013; 193:651 - 69; http://dx.doi.org/10.1534/genetics.112.146704; PMID: 23463798
- Ghosal S, Das S, Chakrabarti J. Long noncoding RNAs: new players in the molecular mechanism for maintenance and differentiation of pluripotent stem cells. Stem Cells Dev 2013; 22:2240 - 53; http://dx.doi.org/10.1089/scd.2013.0014; PMID: 23528033
- Qiu MT, Hu JW, Yin R, Xu L. Long noncoding RNA: an emerging paradigm of cancer research. Tumour Biol 2013; 34:613 - 20; http://dx.doi.org/10.1007/s13277-013-0658-6; PMID: 23359273
- Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D. Ultraconserved elements in the human genome. Science 2004; 304:1321 - 5; http://dx.doi.org/10.1126/science.1098119; PMID: 15131266
- Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, Fabbri M, Cimmino A, Lee EJ, Wojcik SE, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell 2007; 12:215 - 29; http://dx.doi.org/10.1016/j.ccr.2007.07.027; PMID: 17785203
- Mestdagh P, Fredlund E, Pattyn F, Rihani A, Van Maerken T, Vermeulen J, Kumps C, Menten B, De Preter K, Schramm A, et al. An integrative genomics screen uncovers ncRNA T-UCR functions in neuroblastoma tumours. Oncogene 2010; 29:3583 - 92; http://dx.doi.org/10.1038/onc.2010.106; PMID: 20383195
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet 2011; 12:861 - 74; http://dx.doi.org/10.1038/nrg3074; PMID: 22094949
- Lujambio A, Portela A, Liz J, Melo SA, Rossi S, Spizzo R, Croce CM, Calin GA, Esteller M. CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultraconserved regions in human cancer. Oncogene 2010; 29:6390 - 401; http://dx.doi.org/10.1038/onc.2010.361; PMID: 20802525
- Lujambio A, Esteller M. How epigenetics can explain human metastasis: a new role for microRNAs. Cell Cycle 2009; 8:377 - 82; http://dx.doi.org/10.4161/cc.8.3.7526; PMID: 19177007
- Scaruffi P, Stigliani S, Moretti S, Coco S, De Vecchi C, Valdora F, Garaventa A, Bonassi S, Tonini GP. Transcribed-Ultra Conserved Region expression is associated with outcome in high-risk neuroblastoma. BMC Cancer 2009; 9:441; http://dx.doi.org/10.1186/1471-2407-9-441; PMID: 20003513
- Jones PA, Baylin SB. The epigenomics of cancer. Cell 2007; 128:683 - 92; http://dx.doi.org/10.1016/j.cell.2007.01.029; PMID: 17320506
- Lujambio A, Esteller M. CpG island hypermethylation of tumor suppressor microRNAs in human cancer. Cell Cycle 2007; 6:1455 - 9; http://dx.doi.org/10.4161/cc.6.12.4408; PMID: 17581274
- Hudson RS, Yi M, Volfovsky N, Prueitt RL, Esposito D, Volinia S, Liu CG, Schetter AJ, Van Roosbroeck K, Stephens RM, et al. Transcription signatures encoded by ultraconserved genomic regions in human prostate cancer. Mol Cancer 2013; 12:13; http://dx.doi.org/10.1186/1476-4598-12-13; PMID: 23409773
- Braconi C, Valeri N, Kogure T, Gasparini P, Huang N, Nuovo GJ, Terracciano L, Croce CM, Patel T. Expression and functional role of a transcribed noncoding RNA with an ultraconserved element in hepatocellular carcinoma. Proc Natl Acad Sci U S A 2011; 108:786 - 91; http://dx.doi.org/10.1073/pnas.1011098108; PMID: 21187392
- Sana J, Hankeova S, Svoboda M, Kiss I, Vyzula R, Slaby O. Expression levels of transcribed ultraconserved regions uc.73 and uc.388 are altered in colorectal cancer. Oncology 2012; 82:114 - 8; http://dx.doi.org/10.1159/000336479; PMID: 22328099
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005; 102:15545 - 50; http://dx.doi.org/10.1073/pnas.0506580102; PMID: 16199517
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med 2005; 353:1793 - 801; http://dx.doi.org/10.1056/NEJMoa050995; PMID: 16251535
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006; 9:189 - 98; http://dx.doi.org/10.1016/j.ccr.2006.01.025; PMID: 16530703
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 2006; 103:2257 - 61; http://dx.doi.org/10.1073/pnas.0510565103; PMID: 16461460
- Fabbri M, Garzon R, Andreeff M, Kantarjian HM, Garcia-Manero G, Calin GA. MicroRNAs and noncoding RNAs in hematological malignancies: molecular, clinical and therapeutic implications. Leukemia 2008; 22:1095 - 105; http://dx.doi.org/10.1038/leu.2008.30; PMID: 18323801
- Ma L, Reinhardt F, Pan E, Soutschek J, Bhat B, Marcusson EG, Teruya-Feldstein J, Bell GW, Weinberg RA. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol 2010; 28:341 - 7; http://dx.doi.org/10.1038/nbt.1618; PMID: 20351690
- Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjärn M, Hansen HF, Berger U, et al. LNA-mediated microRNA silencing in non-human primates. Nature 2008; 452:896 - 9; http://dx.doi.org/10.1038/nature06783; PMID: 18368051
- Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G 3rd, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med 2008; 14:723 - 30; http://dx.doi.org/10.1038/nm1784; PMID: 18587408
- Costa FF. Non-coding RNAs and new opportunities for the private sector. Drug Discov Today 2009; 14:446 - 52; http://dx.doi.org/10.1016/j.drudis.2009.01.008; PMID: 19429503
- Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol 2010; 12:247 - 56; PMID: 20173740
- Rodríguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med 2011; 17:330 - 9; http://dx.doi.org/10.1038/nm.2305; PMID: 21386836
- Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 2009; 137:1005 - 17; http://dx.doi.org/10.1016/j.cell.2009.04.021; PMID: 19524505