Abstract
Post-transcriptional cleavage of RNA molecules to generate smaller fragments is a widespread mechanism that enlarges the structural and functional complexity of cellular RNomes. Substrates for such RNA fragmentations are coding as well as non-protein-coding RNAs. In particular, fragments derived from both precursor and mature tRNAs represent one of the rapidly growing classes of post-transcriptional RNA pieces. Importantly, these tRNA fragments possess distinct expression patterns, abundance, cellular localizations, or biological roles compared with their parental tRNA molecules. Here we review recent reports on tRNA cleavage and attempt to categorize tRNA pieces according to their origin and cellular function. The biological scope of tRNA-derived fragments ranges from translation control, over RNA silencing, to regulating apoptosis, and thus clearly enlarges the functional repertoire of ncRNA biology.
Introduction
Our functional understanding of non-protein-coding RNA (ncRNA) biology has undergone dramatic changes in the recent past. With the dawn of the new millennium, initial Sanger sequencing studies challenged the common view that cellular life relies basically only on three main transcript classes, namely on messenger RNAs (mRNAs), transfer RNAs (tRNAs), and ribosomal RNAs (rRNAs). These pioneer studies revealed hundreds of additional ncRNA candidates in all three domains of life, thus indicating a so far hidden layer of transcriptome complexity.Citation1,Citation2 With the power of high-throughput sequencing technology, more comprehensive insight into whole transcriptomes became feasible and confirmed that we have thus far only scratched the tip of the ncRNA iceberg. It was realized that genomes of higher eukaryotes are almost entirely transcribed into RNA while only a minor portion thereof was subsequently translated into proteins (reviewed in refs. Citation3 and Citation4). These findings suggested a very intricate ncRNA-based network of gene regulation and led to picture “the eukaryotic genome as an RNA machine.”Citation5 While initially small ncRNAs have been primarily implicated in post-transcriptional gene silencing by targeting mRNAs, more recent evidence strongly suggest that they can also act on DNA by playing pivotal roles in regulating genome stability and guiding chromatin remodeling (reviewed in ref. Citation6). The latest dramatic change in our understanding of the complexity of cellular RNomes arose with the demonstration that ncRNA transcripts with well-described functions, such as small nucleolar RNAs, can serve as precursors for downstream cleavage events generating yet another class of functional RNA pieces, namely microRNAs (miRNAs).Citation7,Citation8 Subsequent studies in a variety of model organisms revealed that such post-transcriptional RNA fragmentation into smaller entities is widespread and likely conserved.Citation9,Citation10 RNA cleavage involves all sorts of transcripts, including coding and non-protein-coding RNAs, thus significantly diversifying the complexity and functional repertoire of the transcriptome. Here, we review recent efforts to understand post-transcriptional tRNA fragmentation and we attempt to link the different classes of tRNA-derived pieces to particular cellular functions.
Functions of Genuine tRNA
tRNAs are universally conserved key components of the translational machinery, linking the genetic code with the amino acid sequence of proteins. tRNAs have a length of 73–90 nucleotides (nt) and have a characteristic secondary and tertiary structure. The backbone of the so-called cloverleaf structure is made of four base-paired stems and five unpaired regions: the D-loop, anticodon loop, variable loop, T-loop, and in case of mature tRNAs, the 3′ CCA tail (). For its adaptor function, each tRNA is charged with an amino acid that is covalently linked to the adenosine of the 3′CCA end, a reaction that is performed by cognate aminoacyl-tRNA synthetases. By reading the mRNA three nucleotides at a time by base-paring, the codon (mRNA)–antidocon (tRNA) interaction defines the amino acid position in the protein.Citation11 On the ribosome, tRNAs do not solely serve as passive substrate for amino acid polymerization, but provide a functional group (the ribose 2’ hydroxyl at the 3′ terminal adenosine) that has been proposed to directly participate in the chemistry of peptide bond formation.Citation12-Citation15
Figure 1. Processing and function of tRNA pieces. Precursor tRNA (pre-tRNA) transcripts are processed by RNase P, RNase Z, and the splicing endonuclease to remove the 5′ leader, 3′ trailer, and (if present) intronic sequences (gray), respectively. If the 3′ CCA end is not encoded, the CCA-adding enzyme finally generates the crucial 3′ end, thus resulting in mature tRNAs. Both pre-tRNA and mature tRNAs can give rise to smaller tRNA pieces. Depending on their origin, they are referred to as 5′ leader-exon tRF, 3′ U tRF, 5′ tRF, 3′ CCA tRF, or as 5′ and 3′ tRNA halves (red). Diverse nucleases identified or suggested to be involved in tRNA maturation and tRNA fragmentation are listed in blue. Whereas the endonucleases involved in tRNA halves production are well studied (PrrC, colicin D, and colicin E5 in bacteria; Rny1 and γ-toxin in certain yeast strains; angiogenin in human), the processing enzymes involved in tRF generation are less clear (indicated by dashed arrows and question marks). Potential cellular targets of the individual tRNA fragment classes are indicated (purple box) and the suggested functions are given (yellow box). Functional evidence for some tRNA pieces is less clear and this is indicated by dashed lines. References for the functional implications of tRNA-derived pieces are listed at the bottom.
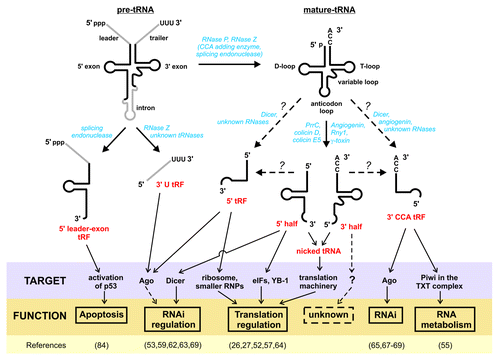
Beside this well-known canonical role during protein biosynthesis, tRNAs have been shown to carry out additional functions, since they are involved in the regulation of numerous metabolic and cellular processes, in both prokaryotes and eukaryotes. In E. coli for example, uncharged tRNAs act as signaling molecules under nutritional stress, activating the stringent response pathway and thereby promoting cell survival.Citation16 A similar function in gene expression has been described in yeast and human cells, where tRNAs can activate Gcn2 kinase, which modulates amino acid starvation response (reviewed in ref. Citation16). Uncharged tRNAs can further act as a substrate for a riboswitch-like gene regulation, the so called T-box mechanism, where they serve as sensors for amino acid deprivation. Upon binding to T-box elements located in the 5′ UTR of cistronic bacterial mRNAs, they are capable of regulating aminoacyl-tRNA synthetase genes or genes involved in amino acid biosynthesis and uptake.Citation17 Besides acting as stress-sensors, tRNAs can serve as primers for the replication of viruses, participate in cell wall synthesis, and in the biosynthesis of certain porphyrines and amino acids (reviewed in ref. Citation11). Given this functional repertoire of full-length tRNA molecules, it is thus not astonishing that they represent one of the most abundant cellular transcripts and amount up to 15% of the total cellular RNAs.Citation16 Due to their universally conserved L-shaped three-dimensional conformation, which is firmly stabilized by extensive secondary and tertiary structural contacts, tRNA molecules belong to the most stable RNAs in a cell.Citation18 With half lives of hours to days, tRNAs are more robust than mRNAs, which merely persist for minutes to hours.Citation19 Folding and stability of tRNA molecules is further controlled by numerous post-transcriptional modifications that cluster primarily in the anticodon-stem loop, the D- and T-stems.Citation20 Since structural and functional integrity of tRNA molecules is so crucial for proper cellular functioning, many control pathways evolved, which recognize and degrade misfolded or hypomodified tRNA molecules rapidly (reviewed in ref. Citation21). It is therefore not surprising that for a long time stable tRNA cleavage intermediates have been regarded as useless debris, and were thus overlooked and omitted from further functional analysis.
tRNA in Pieces
Already in the 1990s, Levitz and Chapman observed that certain E. coli strains carrying a plasmid-encoded PrrC endonuclease are able to target tRNAs for cleavage within the anticodon loop in response to T4 phage infection.Citation22 By depleting its own tRNALys pool, the infected cell is able to reduce translation of late T4 proteins and therefore interferes with virus propagation. Nevertheless, even after the deep-sequencing revolution, tRNA pieces were not appreciated as functional entities and were thus considered as random degradation products or sequencing artifacts. This view changed with the observation, that already known functional ncRNAs are targets for further downstream processing and can give rise to even smaller RNA entities with novel cellular roles (reviewed in ref. Citation23). Among these processed RNA fragments are tRNA-derived pieces that represent a rapidly growing class of novel regulatory ncRNAs. This class of riboregulators is, however, quite heterogeneous in two respects: first, the cleavage of tRNAs can occur at different sites and involves different tRNA isoacceptors, depends on the cell’s environment, and can change during development. Second, the functions of the tRNA-derived pieces proposed so far do not yet allow drawing a clear-cut picture of its ncRNA biology. Therefore, the following sections aim to provide a general overview of what is known so far about tRNA fragments and unravel functional similarities and common principles of tRNA-derived ncRNA pieces. Throughout this review, we follow the proposed nomenclature by Hutvagner and colleagues.Citation24
tRNA halves
tRNA halves (also known as “tRNA-derived, stress-induced small RNAs” or short tiRNAs),Citation25,Citation26 deriving from both the 5′ and 3′ part of full-length tRNA, are produced by cleavage in the anticodon loop (). It is important to note that the targeted tRNAs are typically fully matured tRNA transcripts, carrying an entire set of nucleoside modifications, lacking introns, and have processed 5′ and 3′ termini, often including the 3′ CCA sequences.Citation27,Citation28 The trigger for tRNA cleavage is in the majority of cases nutritional, biological, or physicochemical stress, although tRNA halves have also been observed under unstressed conditions.Citation27,Citation29 Furthermore, the extent of cleavage depends on the nature of stress.Citation30 For example, in Tetrahymena thermophila tRNA halves production has been detected under cold shock, but not during heat shock.Citation31 Importantly, human tRNAs do not respond to apoptosis-inducing agents like γ-irradiation,Citation28 etoposide-treatment,Citation25 or caffeine treatment,Citation25,Citation32 indicating cell death-independent pathways for tRNA cleavage. The best described enzymes responsible for cleavage are Rny1Citation33 in yeast and angiogeninCitation32 in higher eukaryotes (). Rny1 is a member of the RNase T2 family and its human ortholog is RNASET2. Both act as sensors of cellular damage and can promote cell death and inhibit tumor formation, independent of their nuclease activity. However, RNASET2 does not affect tRNA cleavage in mammalian cells.Citation33 Angiogenin, a member of the RNase A family, has already been described in 1985 as an angiogenic protein that was secreted into conditioned media of cultured adenocarcinoma cells, especially under hypoxic conditions.Citation34 Beside its function in blood vessel growth and related tumor progression,Citation35 it became evident that angiogenin is a cytotoxic tRNA-specific ribonuclease.Citation36 Under normal conditions, both ribonucleases, Rny1 and angiogenin, are translocated into the nucleus but are released into the cytoplasm under certain stress conditions. However, cleavage by angiogenin can be regulated by tRNA methylation mediated by the DNA methyltransferase Dnmt2Citation37 or by the ribonuclease inhibitor RNH1.Citation38 Similar mechanisms of tRNA halves production have been described in some bacterial species, where bacterial tRNA anticodon nucleases like PrrC, colicin D, and colicin E5, cleave specific subsets of tRNAs in response to invading species or stress-induced signals, thereby functioning as a kind of innate immune system (reviewed in ref. Citation39). The γ-toxin in the killer strains of the dairy yeast Kluyveromyces lactis is another described tRNA anticodon nuclease. When secreted, this toxin causes irreversible arrest of sensitive yeast cells in the G1 phase of the cell cycle, which is caused by targeting certain tRNAs for cleavage in the anticodon region.Citation40
Conspicuously, two different kinds of cleavage phenomena have been observed. While global tRNA cleavage correlated with codon usage in organisms like Streptomyces coelicolor,Citation41,Citation42 other groups reported on isoacceptor-specific preferences. Here no correlation between targeted tRNAs and their number of genes, their cellular copy number, or the codon usage, has been found.Citation43-Citation45 Since there is no obvious consistency with respect to the preferred isoacceptors targeted for tRNA halves production in different organisms, this process seems to be species-specific. However, in both cleavage scenarios, the stability of the two generated tRNA halves can be asymmetric and change within the same organism, depending on the encountered stress condition and life stage.Citation42,Citation46 This is well exemplified in two independent transcriptome studies in the unicellular eukaryote Trypanosoma cruzi. Franzen et al. detected 88.9% of the tRNA halves deriving from the 3′ part of the genuine tRNA of epimastigotic organisms, with a preference for tRNAHis.Citation47 However, under nutritional stress, 98% of the observed tRNA halves originated from the 5′ part, mainly from tRNAGlu and tRNAAsp.Citation48 This situation changes again in the metacyclic life form of this organism, when primarily 3′ tRNA halves of yet again different isoacceptos have been detected under the same conditions.Citation48 Furthermore, the tRNA halves exhibited differences with respect to their subcellular localization. Whereas they accumulate within posterior cytoplasmic granules of unknown structure and composition in epimastigotes, metacyclics show mainly uniform cytoplasmic distribution.Citation44 These observations of distinct tRNA halves production and localization hint at a functional relevance of tRNA cleavage rather than to a random degradation of tRNAs.
What are the functions of tRNA halves? Since tRNAs are key components of the translational machinery, inhibition of protein production as a consequence of deprivation of full-length tRNAs due to tRNA cleavage appears to be a plausible scenario (). Indeed, soon after angiogenin has been discovered as a human plasma protein, it has been demonstrated to possess cytotoxic tRNA-specific ribonuclease activity that abolished protein synthesis when added to Xenopus oocytes and rabbit reticulocyte lysates. The severity of translation inhibition correlated with the extent of tRNA cleavage.Citation36 A similar effect was seen when yeast tRNAs were first nicked with RNase A in vitro and subsequently used in translation reactions utilizing the wheat germ system.Citation49 At first glance this all makes sense: cleaving certain tRNA molecules negatively affects protein biosynthesis. However, this scenario appears to be too simple, since it has been shown that the steady-state level of genuine tRNAs does not actually change significantly upon tRNA halves production.Citation50,Citation51 Furthermore, tRNA cleavage during stress typically affects only a very small tRNA portion, which is consistently lower than that of the corresponding full-length tRNAs. The simple translation inhibition model is further challenged by the fact that the two tRNA halves are typically unequally stable in vivo. This suggests that the tRNA half molecules might be functional on their own and could fulfill a cellular role independent of their cognate partner half.
While several groups proposed tRNA cleavage as a mechanism to eliminating uncharged or redundant tRNAs,Citation28,Citation41 two publications from the Anderson lab suggest a more intricate role of tRNA halves during translation control. They observed that stress-induced 5′ tRNA halves (but not the respective 3′ halves) were able to inhibit translation and promote stress granule assembly in mammalian cells.Citation26 These granules are mainly composed of stalled pre-initiation complexes, suggesting the translational initiation machinery being targeted by the 5′ tRNA halves. Consistently, they demonstrated that certain 5′ tRNA halves (Ala/Cys), possessing a putative terminal oligo-G motif containing four to five consecutive guanosines, contribute in displacing translation initiation factors from both capped (eIF4E/G/A) and uncapped (eIF4G/A) mRNAs by cooperating with the translational silencer protein YB-1. This complex then activates a cytoprotective stress response, resulting in stress granule formation.Citation52 Notably, stress granules have still been observed even in the case YB-1 was removed by RNAi, thus leaving many mechanistic questions still unanswered how 5′ tRNA halves are involved in translation control. Similar inhibitory effects of 5′ tRNA halves on translation were recently observed in the plant Arabidopsis thaliana,Citation27 but with some notable differences: (1) diverse tRNA pieces, including tRNA halves, have been observed independent of stress and, (2) the inhibitory effect of the 5′ tRNA halves on in vitro translation did not correlate with the number of consecutive guanine residues at the 5′ end as proposed by Anderson et al. Furthermore, it was demonstrated that the human tRNA halves (Ala/Cys), which repressed translation initiation in rabbit reticulocyte lysate,Citation52 showed no inhibitory effect in wheat germ extract. This either hints to species specificity or to missing cofactors that determine the inhibitory potential.
It appears therefore that there is still much to be learned about tRNA halves in translation regulation. The proposed 5′ terminal oligo-G motifCitation52 raises several questions concerning specificity: (1) Is any RNA with a 5′ terminal guanine stretch capable of blocking translation initiation? (2) What is the biological function of all the non-oligo-G-containing tRNA halves and the corresponding 3′ partners? (3) Given that human 5′ tRNA halves do not seem to function in plant translation systems, which co-factors are responsible for the species specificity? And which factors influence the asymmetric stability of the 5′ and 3′ tRNA halves in various organisms?
Very recently evidence has been presented that further enlarges the functional scope of tRNA halves. It was shown that tRNA halves may not only regulate translation, but also small RNA homeostasis during heat-shock.Citation53 Increased tRNA cleavage, induced by decreased tRNA stability (e.g., under certain stress conditions or lacking methylation in the anticodon loop), seems to interfere with the small-interfering RNA (siRNA) pathway through binding of tRNA halves to Dicer-2 (). Consequentially, siRNA production is impaired and allows translation of stress-relevant mRNAs.Citation53
tRNA-derived fragments
Beside tRNA halves, shorter tRNA-derived fragments (tRFs) in the size range of 13–30 nts have been described in organisms spanning all domains of life. Cleavage does not occur randomly, and depending from which part of the pre-tRNA or mature tRNA they are derived, tRFs were classified as 5′ tRFs, 3′ CCA tRFs, 3′ U tRFs, or 5′ leader-exon tRFs (). Since these tRFs differ in terms of origin, processing, and potential function, they will be discussed separately, as follows.
5′ tRFs
5′ tRFs are formed by cleavage in or around the D loop of tRNAs (). As described for tRNA halves, the precursor of these fragments is most likely mature tRNA, since 5′ leader sequences are absent. Therefore, a generation of 5′ tRFs can occur at any point of tRNA processing, given that RNase P has already removed the 5′ leader of the pre-tRNA. Importantly, since many studies report on mixed populations of tRFs and tRNA halves,Citation27,Citation47,Citation54 it cannot be excluded that tRFs are produced from 5′ tRNA halves by 3′ exonuclease chew-back (). Similar to tRNA halves, the abundance of 5′ tRFs does not correlate with the number of the parent gene copies, gene numbers, and codon usage, with the exception of tRFs described in Tetrahymena.Citation55 In contrast, in many cases the majority of observed 5′ tRFs originates from a single isoacceptor tRNA species. Furthermore, differences in length have been reported: in Arabidopsis thaliana roots 80% of tRFs correspond to a 19 nt-long 5′ part of tRNAGly,Citation43 in embryonic calli of rice 82% of 5′ tRFs correspond to a 20–22 nt-long pieces of tRNAAla,Citation56 while in the archaeon Haloferax volcanii 85% of all observed 5′ tRFs originate from a 26 nt-fragment of tRNAVal.Citation57 These differences in tRNA specificities, abundance, and length support the conclusion that distinct biogenesis mechanisms are at work.
The characterized size range of 19–26 nts for 5′ tRFs and the primarily double-stranded nature of the precursors (namely the mature tRNAs) prompted some research groups to look for functional similarities between 5′ tRFs and small RNAs of the RNA interference (RNAi) machinery. This class of small ncRNAs (sized ~20–30 nts) includes members of the miRNA, siRNA, and Piwi-interacting RNA (piRNA) families and are efficient regulators of gene expression and genome stability (reviewed in ref. Citation58). While piRNAs are processed independently of Dicer, all small RNAs of the RNA silencing class share association with a member of the argonaute (Ago) protein family, the key component of the RNA-Induced Silencing Complex (RISC). In HeLa cells, a 19 nt-long 5′ tRF from tRNAGlu showed equal abundance and processing scores compared with genuine miRNAs.Citation59 Processing of this 5′ tRF has been demonstrated to be sensitive to Dicer knockdown, thus implying a mi/siRNA-like behavior of Glu-tRF. However, this interpretation is not necessarily completely conclusive since a possible general tRNA destabilization caused by Dicer knockdown-dependent slow-down of cell growth was not excluded in this study as cause for the observed effects.Citation60 Despite a possible Dicer-processing of this 5′ tRF, it clearly differs from canonical miRNAs in several points: (1) the tested 5′ tRF showed only poor association with Ago proteins and has been identified in fractions smaller than the minimal RISC and (2) the 5′ tRF is modified at the terminal ribose, a feature that is absent in human miRNAs. In fact, terminal modifications of the 5′ tRFs can potentially explain the weak Ago association.Citation10 A Dicer-independent 5′ tRF production has been observed in Schizosaccharomyces pombe.
Furthermore, it has been shown that a complex called TRAMP prevents tRNA and rRNA species from entering the small RNA pathway.Citation61 Therefore, a highly abundant 23 nt-long 5′ tRF of tRNAGlu only showed Ago1 association in a cid14Δ strain, lacking the poly(A)-polymerase of the TRAMP complex that normally targets aberrant RNA for degradation via the nuclear exosome. In contrast, clear Ago binding of 5′ tRFs has been described in human monocytic cells (Ago1)Citation62 and Arabidopsis thaliana (Ago1, 2, 4, and 7).Citation63 It is of note that not all tRFs of the latter study associated with Ago proteins. Given the fact that any abundant small RNA, which is in the size range of si/miRNA, can in principle associate with certain Ago proteinsCitation61 and that even full-length tRNAs co-immunoprecipitate with Ago,Citation59 challenges the biological relevance of the putative tRF-Ago associations. In the canonical RNAi pathway, si/miRNAs target mRNAs by binding to fully or partially complementary regions. Several putative 5′ tRF-targeted mRNAs have been identified by bioinformatic tools;Citation62 however, experimental validation is still missing. A further possible function for 5′ tRFs in the RNAi pathway has been suggested in which 5′ tRFs compete as non-specific small RNA mimics with components of the RNAi machinery. In this scenario, tRFs might impair si/miRNA homeostasis by interfering with si/miRNA processing and RISC loading.Citation18
A markedly different cellular function for 5′ tRFs has been very recently proposed.Citation57 In the archaeon Haloferax volcanii, a 26 nt-long tRF originating from tRNAVal was shown to directly bind to ribosomes in vitro and in vivo, and was capable of inhibiting translation (). This Val-tRF exclusively associates with the small ribosomal subunit especially upon alkaline stress conditions. The Val-tRF does not interfere with tRNA binding to the ribosome but might compete with productive mRNA positioning on the small ribosomal subunit (J Gebetsberger and N Polacek, manuscript in preparation). Support for such a ribosome-targeted role of 5′ tRFs during translation control has been recently obtained in human cells. Hutvagner et al. reported that 5′ tRFs were able to efficiently inhibit in vitro translation, which was independent of mRNA target sites.Citation64 By examining the inhibitory potential of other human 5 ’tRFs, a universally conserved GG dinucleotide at the 3′ terminus has been proposed as crucial functional determinant. Nevertheless, such a “mini motif” does not appear to be sufficient, since one of the identified 5′ tRFs that contained the GG sequence failed to inhibit translation. Even though 5′ tRNA halves also repress protein biosynthesis (see above),Citation52 the mechanisms clearly differ: (1) the proposed terminal oligo-G motif of 5′ tRNA halves is either absent in the investigated 5′ tRFs or not crucial for the inhibitory potential, (2) 5′ tRFs do not target mRNA, (3) whereas 5′ halves associate with translation initiation factors and the translational silencer protein YB-1, 5′ tRFs associate with ribosomal particles (30S ribosomal subunits and polysomes in H. volcanii; polysomes, and RNPs smaller than 40S subunits in human cells).Citation57,Citation64 How exactly these 5′ tRFs perform their action on the ribosome, and how they are processed awaits experimental clarification.
3′ CCA tRFs
3′ CCA tRFs are derived from the 3′end of tRNAs by cleavage in or at the T loop and carry the trinucleotide CCA at the acceptor stem, the characteristic 3′ end of mature tRNAs (). The enzyme responsible for processing is most likely, although not exclusively, Dicer.Citation65-Citation67 Alternatively, angiogenin, the enzyme responsible for the generation of tRNA halves, or other RNase A members, were proposed for Dicer-independent processing.Citation68
Nearly all functionally characterized 3′ CCA tRFs have in common that they associate with components of the RNAi machinery and exert, at least in some cases, miRNA-like functions.Citation67-Citation69 In an analysis of small RNAs in human embryonic kidney cells, Haussecker et al. identified two different groups of tRFs.Citation69 While one of them was composed of Dicer-independent trailer-derived RNAs (termed type II tsRNA; will be discussed later on), they also identified a group of 3′ CCA tRFs (type I tsRNA), whose generation was Dicer-dependent. By associating with Ago1–4, these tRFs showed modest trans-silencing capacities (), similar to that of canonical mi/siRNAs. However, in contrast to canonical small RNAs of the RNAi machinery, these 3′ CCA tRFs showed no association with Mov10, a putative RNA helicase that is part of the RISC. Similar results have been obtained for a 22 nt-long 3′ CCA tRF that is expressed in human B cells.Citation67 The mRNA of the human replication protein RPA1 was identified as bona fide target, a protein that shows essential functions in DNA metabolism, including genome replication, recombination, and DNA repair. Since this 3′ CCA tRF is downregulated in lymphoma cell lines, these findings suggest that corresponding increased levels or RPA1 could result in a growth advantage of malignant cells. Furthermore, evidence has been presented that 3′ CCA tRFs, together with components of the RNAi machinery, establish a mechanism for defending viral infections.Citation65,Citation68 While tRNAs (commonly tRNALys, tRNAPro, and tRNATrp) were shown to serve as primers for the initiation of reverse transcription and viral DNA synthesis,Citation70 Yeung et al. suggested that the duplex formed by binding of the 3′ end of tRNALys to the primer binding site (PBS) of HIV could be, at least in vitro, processed by Dicer.Citation65 They showed that Ago2 association of the resulting 18 nt-long 3′ CCA tRF triggered gene silencing activity in a luciferase reporter assay and that viral replication was modulated upon overexpression or knockdown of this ncRNA. Since similar 3′ CCA tRFs have been shown to be complementary to the replication loci of the human endogenous retrovirus (HERV) and guide Ago2-mediated target cleavage, one could speculate whether these tRFs function as RNAi-based inhibitors of endogenous retroviral replication.Citation68 This is a reasonable hypothesis since the RNAi pathway is thought to have evolved as an immune defense mechanism against invading species in basal organisms that lack protein-based immune systems.Citation71 While all these studies were performed in human cell lines, interesting data also exist for 3′ CCA tRFs in the ciliated protozoan Tetrahymena thermophila.Citation72 In an earlier publication, Couvillion et al. showed that 3′ CCA tRFs are efficiently loaded onto the Piwi protein Twi12.Citation55 While Ago proteins are ubiquitously present in diverse tissues, Piwi proteins are predominantly present in the germ line and associate with piRNAs.Citation73 Twi12 is of particular interest, since this protein is the only Piwi necessary for vegetative growth, lacking slicer activity, and is loaded with Dicer-independent small RNAs. More detailed investigations of the Twi12 protein revealed that it forms a complex with the 5′ to 3′ exoribonuclease Xrn2 and a so far not described protein, which they termed Twi-associated novel 1 (Tan1).Citation72 Interestingly, Xrn2 (Rat1 in yeast) functions in the nucleus and is involved in aberrant mRNA degradation, maturation of rRNA, and in Pol I and Pol II transcription termination.Citation74 Since Xrn2 lacks a nuclear localization signal, the question emerged how the nuclear import is facilitated in the context of the Twi12/Xrn2/Tan1 (TXT) complex. In summary, the authors could show that upon loading with 3′ CCA tRFs, Twi12 forms the TXT complex that targets Xrn2 into the nucleus and stimulates its nuclease activity (). Importantly, 3′ CCA tRFs are required for this process, suggesting that Twi12 senses cytoplasmic tRNA levels and influences ribosome biogenesis upon changes of cellular conditions.Citation75
3′ U tRFs
3′ U tRFs are derived from the 3′end of pre-tRNAs (), commonly starting directly after the 3′end of mature tRNAs and end in a stretch of U residues that are produced by RNA polymerase II transcription run-off.Citation24 Since these trailer sequences are processed through RNase Z cleavage during tRNA maturation, involvement of Dicer seems to be unlikely. However, one study suggested a Dicer-dependent mechanism, where the pre-tRNA is predicted to fold into a bulged hairpin.Citation66 Interestingly, tRNA 3′ trailer sequences are thought to be rapidly degraded in the nuclear compartment soon after being cleaved from pre-tRNAs. Therefore, it is quite surprising that stable 3′ U tRFs are localized mainly in the cytoplasm.Citation76,Citation77 There are two likely explanations: (1) 3′ U tRFs are exported from the nucleus by an unknown mechanism upon processing, or (2) pre-tRNAs escape nuclear quality control and are processed by cytoplasmic 3′ tRNases. Strikingly, there is evidence supporting the second possibility. Based on the observations of pre-tRNA splicing in the cytoplasm, aminoacylation in the nucleus, and retrograde movement of tRNAs, nuclear-cytoplasmic localization of tRNAs appears to be a dynamic and bi-directional process (reviewed in ref. Citation78). Furthermore, there are several reports on cytoplasmic tRNA endonucleases.Citation76,Citation79
In support of this scenario, Lee et al. demonstrated that in human cancer cell lines certain pre-tRNAs are exported from the nucleus and cleaved by the cytoplasmic tRNA endonuclease ELAC2, a homolog of RNase Z.Citation76 Further characterization of one 3′ U tRF, named tRF-1001, showed that it is regulated according to cell proliferation and is necessary for cell viability. Coincidentally, Haussecker et al. analyzed the same 3′ U tRF candidate (called cand45) in the context of investigating their type II tsRNAs (see above).Citation69 Whereas Lee et al. did not investigate Ago association, the other group showed binding to Ago3 and Ago4. While no canonical gene silencing activity could be observed and modulation of 3′ U tRFs levels had only minor effects on miRNA abundance, pronounced changes in silencing activities of mi/siRNAs have been observed. This suggests a 3′ U tRF-guided control of small RNA silencing through differential Ago association, and thus, competition with genuine si/miRNAs. Such a scenario could also explain the effects of 3′ U tRFs on cell proliferation as observed by Lee et al. Since 3′ U tRFs have been detected not only in vertebrates including human, dogs, and chicken,Citation69,Citation76,Citation77,Citation80 but also in archaeaCitation81 that lack the classical RNAi pathway, future studies on the functional similarities across domains and putative cellular role(s) of 3′ U tRFs are eagerly awaited.
Unclassified tRFs
Other, yet unclassified, tRFs do not fit into the general classification scheme regarding their origin and/or length (). We will discuss them shortly, since they expand the functional repertoire of tRNA pieces even further. This class of tRFs includes RNAs of about 46 nt in length in the primitive eukaryote Giardia lamblia that are produced by cleavage of mature tRNAs 5′ of the anticodon loop. The generated 3′ tRFs seem to be the result of global tRNA cleavage, induced by different stress stimuli (e.g., encystation, serum deprivation, cold stress, and heat shock).Citation82 tRFs of aberrant size and yet unknown function have also been detected in other studies, including 3′ quarters of tRNAsCitation27 and “mid”-fragments.Citation83 While most of the so far identified tRFs derive from mature tRNAs (with the exception of 3′U tRFs), Hanada et al. recently reported on novel tRFs originating from aberrant pre-tRNA splicing.Citation84 Mainly arising from tRNATyr, these tRFs comprise the 5′ leader followed by the 5′ exon of pre-tRNA (). Functional analyses demonstrated that this 5′ leader-exon-tRF sensitizes cells to oxidative-stress-induced activation of the p53 tumor suppressor pathway and p53-mediated cell death. Although the molecular mechanism behind this hitherto unknown tRF class is not yet fully understood, it clearly suggests that the functional repertoire of tRNA-derived pieces goes beyond translation regulation and RNA interference.
Perspectives
Application of the new high-throughput sequencing technologies promoted the discovery of novel regulatory ncRNA candidates and expanded our knowledge on the diversity and complexity of the transcriptome. Nevertheless, caution should be taken in categorizing sequencing reads as functional ncRNAs, since these molecules could be of questionable biological relevance. Random RNA degradation, spurious transcription (“transcriptional noise”), or artifacts of the cDNA library preparations should not be excluded as source for an observed RNA sequence read.Citation3 In particular, tRNAs are known to be heavily modified, and thus, represent a hurdle for efficient reverse transcription. This creates two problems: (1) hypermodified tRNA pieces might be underrepresented in cDNA libraries and (2) not all deep sequencing reads deriving from tRNA loci represent true tRNA pieces. Thus, it is crucial to conduct experimental approaches and to assign a cellular function to an RNA candidate, before referring to it as a novel ncRNA species.
Since rRNAs and tRNAs constitute the vast majority of cellular RNA molecules, it would be reasonable that they also generate more “meaningless” degradation products than other RNAs. Therefore, the explosion of recently detected tRNA pieces in various model systems, spanning all domains of life (), can also be seen from this perspective. Nevertheless, the following observation seems to substantiate the view that tRNA pieces represent biological relevant entities rather than mere degradation products of full-length tRNAs: (1) they show characteristic expression patterns across different cell lines and organisms, (2) their production is specifically induced only during certain environmental conditions and (3) does not necessarily correlate with codon usage and/or gene copy number, (4) tRNA cleavage often occurs at specific sites in the tRNA and is more precise than for many miRNAs and, most importantly, (5) the pool of available mature tRNAs is not significantly altered due to tRNA fragmentation.
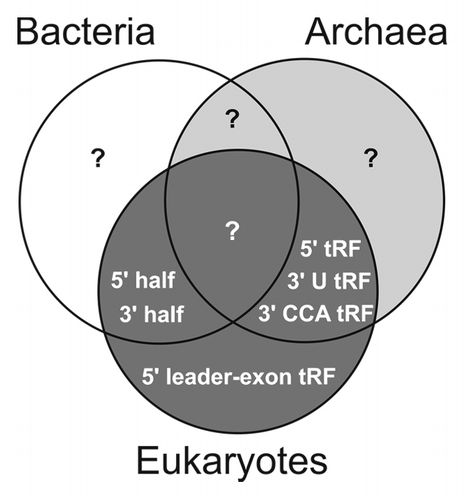
Figure 2. Distribution of tRNA pieces among the three domains of life. A Venn diagram illustrates the verified presence of the various tRNA-derived RNA pieces in prokaryotes (bacteria and archaea) and eukaryotes. Note that the absence of a particular tRNA fragment species in one of the domains does not necessarily imply its absence rather than it reflects our current experimental insight.
However, the functional role of tRFs and tRNA halves is still quite elusive and the biological picture thus incomplete. Although many interesting suggestions on their biological relevance have been made, unequivocal experimental data are often missing. The assumption that cleaved tRNAs can function as canonical miRNAs is one such example. Considering the classical definition of miRNAs, the possibility should not be ruled out that tRFs are more like siblings and not identical twins of miRNAs, since they differ in many aspects from genuine miRNAs, including origin, processing, and Ago association (discussed in ref. Citation60). It is still conceivable that tRFs represent a control mechanism of the silencing machinery, reflected by differential Ago occupancy, and thus, by competing with mi/siRNAs for RISC loading. Considering the TRAMP complex in S. pombe that prevents aberrant RNAs from entering the RNAi pathway, it is reasonable that other organisms could achieve the same goal by titration of silencing components via tRF production.Citation85
It is still unknown which properties determine the tRNAs’ fate of becoming a tRF, and thus, a novel regulatory ncRNA. Mature tRNAs contain more than 100 modified nucleosides and the role of many of them is still elusive.Citation11 Most of them occur non-randomly and cluster in conserved domains. It is worth proposing that some of the tRNA modifications guide RNases, and thus, tRNA processing to specific positions (discussed in refs. Citation18 and Citation38). Even though our current knowledge about tRNA pieces is yet incomplete, they clearly represent a novel class of regulatory ncRNAs with surprising cellular roles in all domains of life (). Future work will reveal how they fit into the ever-growing puzzle of RNA biology.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to Javier Martinez, Ronald Micura, Miriam Koch, Andreas Pircher, and Moritz Niemann for critical reading of the manuscript. Grant support derives from the Swiss National Science Foundation (31003A_143388/1) and the Austrian Science Foundation (Y315) to Polacek N.
References
- Hüttenhofer A, Kiefmann M, Meier-Ewert S, O’Brien J, Lehrach H, Bachellerie JP, Brosius J. RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J 2001; 20:2943 - 53; http://dx.doi.org/10.1093/emboj/20.11.2943; PMID: 11387227
- Hüttenhofer A, Brosius J, Bachellerie JP. RNomics: identification and function of small, non-messenger RNAs. Curr Opin Chem Biol 2002; 6:835 - 43; http://dx.doi.org/10.1016/S1367-5931(02)00397-6; PMID: 12470739
- Hüttenhofer A, Schattner P, Polacek N. Non-coding RNAs: hope or hype?. Trends Genet 2005; 21:289 - 97; http://dx.doi.org/10.1016/j.tig.2005.03.007; PMID: 15851066
- Mattick JS. RNA regulation: a new genetics?. Nat Rev Genet 2004; 5:316 - 23; http://dx.doi.org/10.1038/nrg1321; PMID: 15131654
- Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science 2008; 319:1787 - 9; http://dx.doi.org/10.1126/science.1155472; PMID: 18369136
- Sabin LR, Delás MJ, Hannon GJ. Dogma derailed: the many influences of RNA on the genome. Mol Cell 2013; 49:783 - 94; http://dx.doi.org/10.1016/j.molcel.2013.02.010; PMID: 23473599
- Ender C, Krek A, Friedländer MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol Cell 2008; 32:519 - 28; http://dx.doi.org/10.1016/j.molcel.2008.10.017; PMID: 19026782
- Scott MS, Avolio F, Ono M, Lamond AI, Barton GJ. Human miRNA precursors with box H/ACA snoRNA features. PLoS Comput Biol 2009; 5:e1000507; http://dx.doi.org/10.1371/journal.pcbi.1000507; PMID: 19763159
- Mercer TR, Dinger ME, Bracken CP, Kolle G, Szubert JM, Korbie DJ, Askarian-Amiri ME, Gardiner BB, Goodall GJ, Grimmond SM, et al. Regulated post-transcriptional RNA cleavage diversifies the eukaryotic transcriptome. Genome Res 2010; 20:1639 - 50; http://dx.doi.org/10.1101/gr.112128.110; PMID: 21045082
- Tuck AC, Tollervey D. RNA in pieces. Trends Genet 2011; 27:422 - 32; http://dx.doi.org/10.1016/j.tig.2011.06.001; PMID: 21741109
- Giegé R. Toward a more complete view of tRNA biology. Nat Struct Mol Biol 2008; 15:1007 - 14; http://dx.doi.org/10.1038/nsmb.1498; PMID: 18836497
- Dorner S, Polacek N, Schulmeister U, Panuschka C, Barta A. Molecular aspects of the ribosomal peptidyl transferase. Biochem Soc Trans 2002; 30:1131 - 6; http://dx.doi.org/10.1042/BST0301131; PMID: 12440989
- Lang K, Erlacher M, Wilson DN, Micura R, Polacek N. The role of 23S ribosomal RNA residue A2451 in peptide bond synthesis revealed by atomic mutagenesis. Chem Biol 2008; 15:485 - 92; http://dx.doi.org/10.1016/j.chembiol.2008.03.014; PMID: 18439847
- Zaher HS, Shaw JJ, Strobel SA, Green R. The 2′-OH group of the peptidyl-tRNA stabilizes an active conformation of the ribosomal PTC. EMBO J 2011; 30:2445 - 53; http://dx.doi.org/10.1038/emboj.2011.142; PMID: 21552203
- Trobro S, Aqvist J. Mechanism of peptide bond synthesis on the ribosome. Proc Natl Acad Sci U S A 2005; 102:12395 - 400; http://dx.doi.org/10.1073/pnas.0504043102; PMID: 16116099
- Li Y, Zhou H. tRNAs as regulators in gene expression. Sci China C Life Sci 2009; 52:245 - 52; http://dx.doi.org/10.1007/s11427-009-0039-y; PMID: 19294349
- Green NJ, Grundy FJ, Henkin TM. The T box mechanism: tRNA as a regulatory molecule. FEBS Lett 2010; 584:318 - 24; http://dx.doi.org/10.1016/j.febslet.2009.11.056; PMID: 19932103
- Durdevic Z, Schaefer M. tRNA modifications: necessary for correct tRNA-derived fragments during the recovery from stress?. Bioessays 2013; 35:323 - 7; http://dx.doi.org/10.1002/bies.201200158; PMID: 23315679
- Defoiche J, Zhang Y, Lagneaux L, Pettengell R, Hegedus A, Willems L, Macallan DC. Measurement of ribosomal RNA turnover in vivo by use of deuterium-labeled glucose. Clin Chem 2009; 55:1824 - 33; http://dx.doi.org/10.1373/clinchem.2008.119446; PMID: 19696118
- Motorin Y, Helm M. tRNA stabilization by modified nucleotides. Biochemistry 2010; 49:4934 - 44; http://dx.doi.org/10.1021/bi100408z; PMID: 20459084
- Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev 2010; 24:1832 - 60; http://dx.doi.org/10.1101/gad.1956510; PMID: 20810645
- Levitz R, Chapman D, Amitsur M, Green R, Snyder L, Kaufmann G. The optional E. coli prr locus encodes a latent form of phage T4-induced anticodon nuclease. EMBO J 1990; 9:1383 - 9; PMID: 1691706
- Kawaji H, Nakamura M, Takahashi Y, Sandelin A, Katayama S, Fukuda S, Daub CO, Kai C, Kawai J, Yasuda J, et al. Hidden layers of human small RNAs. BMC Genomics 2008; 9:157; http://dx.doi.org/10.1186/1471-2164-9-157; PMID: 18402656
- Sobala A, Hutvagner G. Transfer RNA-derived fragments: origins, processing, and functions. Wiley Interdiscip Rev RNA 2011; 2:853 - 62; http://dx.doi.org/10.1002/wrna.96; PMID: 21976287
- Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol 2009; 185:35 - 42; http://dx.doi.org/10.1083/jcb.200811106; PMID: 19332886
- Emara MM, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N, Hu GF, Anderson P. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem 2010; 285:10959 - 68; http://dx.doi.org/10.1074/jbc.M109.077560; PMID: 20129916
- Nowacka M, Strozycki PM, Jackowiak P, Hojka-Osinska A, Szymanski M, Figlerowicz M. Identification of stable, high copy number, medium-sized RNA degradation intermediates that accumulate in plants under non-stress conditions. Plant Mol Biol 2013; 83:191 - 204; http://dx.doi.org/10.1007/s11103-013-0079-3; PMID: 23708952
- Fu H, Feng J, Liu Q, Sun F, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett 2009; 583:437 - 42; http://dx.doi.org/10.1016/j.febslet.2008.12.043; PMID: 19114040
- Dhahbi JM, Spindler SR, Atamna H, Yamakawa A, Boffelli D, Mote P, Martin DI. 5′ tRNA halves are present as abundant complexes in serum, concentrated in blood cells, and modulated by aging and calorie restriction. BMC Genomics 2013; 14:298; http://dx.doi.org/10.1186/1471-2164-14-298; PMID: 23638709
- Thompson DM, Parker R. Stressing out over tRNA cleavage. Cell 2009; 138:215 - 9; http://dx.doi.org/10.1016/j.cell.2009.07.001; PMID: 19632169
- Andersen KL, Collins K. Several RNase T2 enzymes function in induced tRNA and rRNA turnover in the ciliate Tetrahymena. Mol Biol Cell 2012; 23:36 - 44; http://dx.doi.org/10.1091/mbc.E11-08-0689; PMID: 22049026
- Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 2008; 14:2095 - 103; http://dx.doi.org/10.1261/rna.1232808; PMID: 18719243
- Thompson DM, Parker R. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J Cell Biol 2009; 185:43 - 50; http://dx.doi.org/10.1083/jcb.200811119; PMID: 19332891
- Fett JW, Strydom DJ, Lobb RR, Alderman EM, Bethune JL, Riordan JF, Vallee BL. Isolation and characterization of angiogenin, an angiogenic protein from human carcinoma cells. Biochemistry 1985; 24:5480 - 6; http://dx.doi.org/10.1021/bi00341a030; PMID: 4074709
- Zhao H, Bojanowski K, Ingber DE, Panigrahy D, Pepper MS, Montesano R, Shing Y. New role for tRNA and its fragment purified from human urinary bladder carcinoma conditioned medium: inhibition of endothelial cell growth. J Cell Biochem 1999; 76:109 - 17; http://dx.doi.org/10.1002/(SICI)1097-4644(20000101)76:1<109::AID-JCB11>3.0.CO;2-K; PMID: 10581005
- Saxena SK, Rybak SM, Davey RT Jr., Youle RJ, Ackerman EJ. Angiogenin is a cytotoxic, tRNA-specific ribonuclease in the RNase A superfamily. J Biol Chem 1992; 267:21982 - 6; PMID: 1400510
- Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev 2010; 24:1590 - 5; http://dx.doi.org/10.1101/gad.586710; PMID: 20679393
- Nawrot B, Sochacka E, Düchler M. tRNA structural and functional changes induced by oxidative stress. Cell Mol Life Sci 2011; 68:4023 - 32; http://dx.doi.org/10.1007/s00018-011-0773-8; PMID: 21833586
- Kaufmann G. Anticodon nucleases. Trends Biochem Sci 2000; 25:70 - 4; http://dx.doi.org/10.1016/S0968-0004(99)01525-X; PMID: 10664586
- Lu J, Esberg A, Huang B, Byström AS. Kluyveromyces lactis gamma-toxin, a ribonuclease that recognizes the anticodon stem loop of tRNA. Nucleic Acids Res 2008; 36:1072 - 80; http://dx.doi.org/10.1093/nar/gkm1121; PMID: 18096622
- Lee SR, Collins K. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J Biol Chem 2005; 280:42744 - 9; http://dx.doi.org/10.1074/jbc.M510356200; PMID: 16272149
- Haiser HJ, Karginov FV, Hannon GJ, Elliot MA. Developmentally regulated cleavage of tRNAs in the bacterium Streptomyces coelicolor. Nucleic Acids Res 2008; 36:732 - 41; http://dx.doi.org/10.1093/nar/gkm1096; PMID: 18084030
- Hsieh LC, Lin SI, Shih AC, Chen JW, Lin WY, Tseng CY, Li WH, Chiou TJ. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol 2009; 151:2120 - 32; http://dx.doi.org/10.1104/pp.109.147280; PMID: 19854858
- Reifur L, Garcia-Silva MR, Poubel SB, Alves LR, Arauco P, Buiar DK, Goldenberg S, Cayota A, Dallagiovanna B. Distinct subcellular localization of tRNA-derived fragments in the infective metacyclic forms of Trypanosoma cruzi. Mem Inst Oswaldo Cruz 2012; 107:816 - 9; http://dx.doi.org/10.1590/S0074-02762012000600018; PMID: 22990974
- Garcia-Silva MR, Cabrera-Cabrera F, Güida MC. A. C. Hints of tRNA-Derived Small RNAs in RNA Silencing Mechanisms. Genes 2012:603-614.
- Zywicki M, Bakowska-Zywicka K, Polacek N. Revealing stable processing products from ribosome-associated small RNAs by deep-sequencing data analysis. Nucleic Acids Res 2012; 40:4013 - 24; http://dx.doi.org/10.1093/nar/gks020; PMID: 22266655
- Franzén O, Arner E, Ferella M, Nilsson D, Respuela P, Carninci P, Hayashizaki Y, Aslund L, Andersson B, Daub CO. The short non-coding transcriptome of the protozoan parasite Trypanosoma cruzi. PLoS Negl Trop Dis 2011; 5:e1283; http://dx.doi.org/10.1371/journal.pntd.0001283; PMID: 21912713
- Garcia-Silva MR, Frugier M, Tosar JP, Correa-Dominguez A, Ronalte-Alves L, Parodi-Talice A, Rovira C, Robello C, Goldenberg S, Cayota A. A population of tRNA-derived small RNAs is actively produced in Trypanosoma cruzi and recruited to specific cytoplasmic granules. Mol Biochem Parasitol 2010; 171:64 - 73; http://dx.doi.org/10.1016/j.molbiopara.2010.02.003; PMID: 20156490
- Zhang S, Sun L, Kragler F. The phloem-delivered RNA pool contains small noncoding RNAs and interferes with translation. Plant Physiol 2009; 150:378 - 87; http://dx.doi.org/10.1104/pp.108.134767; PMID: 19261735
- Saikia M, Krokowski D, Guan BJ, Ivanov P, Parisien M, Hu GF, Anderson P, Pan T, Hatzoglou M. Genome-wide identification and quantitative analysis of cleaved tRNA fragments induced by cellular stress. J Biol Chem 2012; 287:42708 - 25; http://dx.doi.org/10.1074/jbc.M112.371799; PMID: 23086926
- Jöchl C, Rederstorff M, Hertel J, Stadler PF, Hofacker IL, Schrettl M, Haas H, Hüttenhofer A. Small ncRNA transcriptome analysis from Aspergillus fumigatus suggests a novel mechanism for regulation of protein synthesis. Nucleic Acids Res 2008; 36:2677 - 89; http://dx.doi.org/10.1093/nar/gkn123; PMID: 18346967
- Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell 2011; 43:613 - 23; http://dx.doi.org/10.1016/j.molcel.2011.06.022; PMID: 21855800
- Durdevic Z, Mobin MB, Hanna K, Lyko F, Schaefer M. The RNA methyltransferase Dnmt2 is required for efficient Dicer-2-dependent siRNA pathway activity in Drosophila. Cell Rep 2013; 4:931 - 7; http://dx.doi.org/10.1016/j.celrep.2013.07.046; PMID: 24012760
- Wang L, Yu X, Wang H, Lu YZ, de Ruiter M, Prins M, He YK. A novel class of heat-responsive small RNAs derived from the chloroplast genome of Chinese cabbage (Brassica rapa). BMC Genomics 2011; 12:289; http://dx.doi.org/10.1186/1471-2164-12-289; PMID: 21639890
- Couvillion MT, Sachidanandam R, Collins K. A growth-essential Tetrahymena Piwi protein carries tRNA fragment cargo. Genes Dev 2010; 24:2742 - 7; http://dx.doi.org/10.1101/gad.1996210; PMID: 21106669
- Chen CJ, liu Q, Zhang YC, Qu LH, Chen YQ, Gautheret D. Genome-wide discovery and analysis of microRNAs and other small RNAs from rice embryogenic callus. RNA Biol 2011; 8:538 - 47; http://dx.doi.org/10.4161/rna.8.3.15199; PMID: 21525786
- Gebetsberger J, Zywicki M, Künzi A, Polacek N. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea 2012; 2012:260909; http://dx.doi.org/10.1155/2012/260909; PMID: 23326205
- Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature 2009; 457:413 - 20; http://dx.doi.org/10.1038/nature07756; PMID: 19158787
- Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, Green PJ, Barton GJ, Hutvagner G. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 2009; 15:2147 - 60; http://dx.doi.org/10.1261/rna.1738409; PMID: 19850906
- Pederson T. Regulatory RNAs derived from transfer RNA?. RNA 2010; 16:1865 - 9; http://dx.doi.org/10.1261/rna.2266510; PMID: 20719919
- Bühler M, Spies N, Bartel DP, Moazed D. TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat Struct Mol Biol 2008; 15:1015 - 23; http://dx.doi.org/10.1038/nsmb.1481; PMID: 18776903
- Burroughs AM, Ando Y, de Hoon MJ, Tomaru Y, Suzuki H, Hayashizaki Y, Daub CO. Deep-sequencing of human Argonaute-associated small RNAs provides insight into miRNA sorting and reveals Argonaute association with RNA fragments of diverse origin. RNA Biol 2011; 8:158 - 77; http://dx.doi.org/10.4161/rna.8.1.14300; PMID: 21282978
- Loss-Morais G, Waterhouse PM, Margis R. Description of plant tRNA-derived RNA fragments (tRFs) associated with argonaute and identification of their putative targets. Biol Direct 2013; 8:6; http://dx.doi.org/10.1186/1745-6150-8-6; PMID: 23402430
- Sobala A, Hutvagner G. Small RNAs derived from the 5′ end of tRNA can inhibit protein translation in human cells. RNA Biol 2013; 10:553 - 63; http://dx.doi.org/10.4161/rna.24285; PMID: 23563448
- Yeung ML, Bennasser Y, Watashi K, Le SY, Houzet L, Jeang KT. Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res 2009; 37:6575 - 86; http://dx.doi.org/10.1093/nar/gkp707; PMID: 19729508
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev 2008; 22:2773 - 85; http://dx.doi.org/10.1101/gad.1705308; PMID: 18923076
- Maute RL, Schneider C, Sumazin P, Holmes A, Califano A, Basso K, Dalla-Favera R. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci U S A 2013; 110:1404 - 9; http://dx.doi.org/10.1073/pnas.1206761110; PMID: 23297232
- Li Z, Ender C, Meister G, Moore PS, Chang Y, John B. Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs. Nucleic Acids Res 2012; 40:6787 - 99; http://dx.doi.org/10.1093/nar/gks307; PMID: 22492706
- Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 2010; 16:673 - 95; http://dx.doi.org/10.1261/rna.2000810; PMID: 20181738
- Isel C, Ehresmann C, Keith G, Ehresmann B, Marquet R. Initiation of reverse transcription of HIV-1: secondary structure of the HIV-1 RNA/tRNA(3Lys) (template/primer). J Mol Biol 1995; 247:236 - 50; http://dx.doi.org/10.1006/jmbi.1994.0136; PMID: 7707372
- van Rij RP, Saleh MC, Berry B, Foo C, Houk A, Antoniewski C, Andino R. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev 2006; 20:2985 - 95; http://dx.doi.org/10.1101/gad.1482006; PMID: 17079687
- Couvillion MT, Bounova G, Purdom E, Speed TP, Collins K. A Tetrahymena Piwi bound to mature tRNA 3′ fragments activates the exonuclease Xrn2 for RNA processing in the nucleus. Mol Cell 2012; 48:509 - 20; http://dx.doi.org/10.1016/j.molcel.2012.09.010; PMID: 23084833
- Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu Rev Cell Dev Biol 2009; 25:355 - 76; http://dx.doi.org/10.1146/annurev.cellbio.24.110707.175327; PMID: 19575643
- Houseley J, Tollervey D. The many pathways of RNA degradation. Cell 2009; 136:763 - 76; http://dx.doi.org/10.1016/j.cell.2009.01.019; PMID: 19239894
- Hasler D, Meister G. An argonaute protein directs nuclear Xrn2 function. Mol Cell 2012; 48:485 - 6; http://dx.doi.org/10.1016/j.molcel.2012.11.015; PMID: 23200120
- Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev 2009; 23:2639 - 49; http://dx.doi.org/10.1101/gad.1837609; PMID: 19933153
- Liao JY, Ma LM, Guo YH, Zhang YC, Zhou H, Shao P, Chen YQ, Qu LH. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3′ trailers. PLoS One 2010; 5:e10563; http://dx.doi.org/10.1371/journal.pone.0010563; PMID: 20498841
- Hopper AK, Shaheen HH. A decade of surprises for tRNA nuclear-cytoplasmic dynamics. Trends Cell Biol 2008; 18:98 - 104; http://dx.doi.org/10.1016/j.tcb.2008.01.001; PMID: 18262788
- Elbarbary RA, Takaku H, Uchiumi N, Tamiya H, Abe M, Nishida H, Nashimoto M. Human cytosolic tRNase ZL can downregulate gene expression through miRNA. FEBS Lett 2009; 583:3241 - 6; http://dx.doi.org/10.1016/j.febslet.2009.09.015; PMID: 19751732
- Morin RD, O’Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res 2008; 18:610 - 21; http://dx.doi.org/10.1101/gr.7179508; PMID: 18285502
- Heyer R, Dörr M, Jellen-Ritter A, Späth B, Babski J, Jaschinski K, Soppa J, Marchfelder A. High throughput sequencing reveals a plethora of small RNAs including tRNA derived fragments in Haloferax volcanii. RNA Biol 2012; 9:1011 - 8; http://dx.doi.org/10.4161/rna.20826; PMID: 22767255
- Li Y, Luo J, Zhou H, Liao JY, Ma LM, Chen YQ, Qu LH. Stress-induced tRNA-derived RNAs: a novel class of small RNAs in the primitive eukaryote Giardia lamblia. Nucleic Acids Res 2008; 36:6048 - 55; http://dx.doi.org/10.1093/nar/gkn596; PMID: 18820301
- Müller S, Windhof IM, Maximov V, Jurkowski T, Jeltsch A, Förstner KU, Sharma CM, Gräf R, Nellen W. Target recognition, RNA methylation activity and transcriptional regulation of the Dictyostelium discoideum Dnmt2-homologue (DnmA). Nucleic Acids Res 2013; 41:8615 - 27; http://dx.doi.org/10.1093/nar/gkt634; PMID: 23877245
- Hanada T, Weitzer S, Mair B, Bernreuther C, Wainger BJ, Ichida J, Hanada R, Orthofer M, Cronin SJ, Komnenovic V, et al. CLP1 links tRNA metabolism to progressive motor-neuron loss. Nature 2013; 495:474 - 80; http://dx.doi.org/10.1038/nature11923; PMID: 23474986
- Aalto AP, Pasquinelli AE. Small non-coding RNAs mount a silent revolution in gene expression. Curr Opin Cell Biol 2012; 24:333 - 40; http://dx.doi.org/10.1016/j.ceb.2012.03.006; PMID: 22464106