Abstract
Generating heat and maintaining body temperature is the primary function of brown adipose tissue (BAT). Previous studies have implicated microRNAs, including miR-193b and miR-365-1, in BAT differentiation. We used mouse genetics to further understand the specific contributions of these two miRs. BAT function in mice with an inactivated miR-193b-365-1 locus, as determined by their response to the selective β3 adrenergic receptor agonist CL316.243 and their tolerance to cold exposure, was normal and expression of genes associated with functional BAT, including Prdm16 and Ucp1, was unaffected. In addition, genome-wide expression profiles of miRNAs and mRNAs in BAT in the presence and absence of miR-193b-365-1 were determined. In summary, these data demonstrate, in contrast to earlier work, that the development, differentiation, and function of BAT do not require the presence of miR-193b and miR-365-1.
Keywords: :
Introduction
Experimental data suggest that miRNAs can modulate mRNA stability and thereby control the concentration of the corresponding proteins.Citation1,Citation2 Although a large number of studies using either knockdown or overexpression approaches have demonstrated modulatory roles of miRNAs in tissue culture settings in vitro, only a few genetic studies in mice have been published and they provide a less definitive picture.Citation3,Citation4 Adaptive compensatory genetic changes are often invoked in explaining the apparent lack of physiological changes observed upon the disruption of miRNA genes in mice.
The absence of any significant role of miRNAs in BAT development was proposed based on the conditional deletion of Dicer using aP2-Cre transgenic mice.Citation5 However, recent reports proposed a role for some miRNAs in the differentiation and function of brown adipose tissue (BAT).Citation6-Citation8 In addition, a role for miR-133a in the browning of subcutaneous white adipose tissue (SAT) was reported.Citation9 Brown fat is a specialized form of adipose tissue that is critical for non-shivering thermogenesis. Brown adipose cells are derived from MYF5-positive myogenic progenitors and the transcription factor PRDM16 is considered a master regulator that controls the switch between skeletal myoblasts and brown fat cells and also controls thermogenic programs in white adipose tissue.Citation10 The conclusion that specific miRNAs control the function of BAT is frequently based on in vitro differentiation experiments of primary BAT cells from which miRNA functions were ablated using locked nucleic acids (LNA) or in which miRNAs were overexpressed. Specifically, the presence of miR-193b was shown to be critical for the differentiation of primary BAT cells and the expression of key BAT markers, such as UCP1.Citation6 Here we asked whether miR-193b is required for the development of BAT in vivo. The miR-193b-365-1 locus was inactivated in the mouse and physiological and molecular consequences on mRNA and miRNA expression patterns, as well as BAT development and function were determined. In particular, the role of these miRNAs in the compensatory induction of other miRNAs was also investigated.
Results
Inactivation of the miR-193b-365-1 locus
The mir-193b-365-1 locus spans approximately 25 kbp and is located on chromosome 16, between the Mkl2 and Parn genes (). Using ENCODE data we identified two distinct H3K4me3 marks upstream of miR-193b sequences in several cell types, indicating the presence of two specific promoter/regulatory regions (). By gene targeting, mice were generated carrying loxP sites 5′ of the proximal H3K4me3 marks and 3′ of the mature miR-193b sequence (; Fig. S1A). Using a female-specific germline cre deleter mouse strain,Citation11 we generated mice from which 3.1 kbp of DNA spanning the promoter/regulatory region of the miR-193b-365-1 locus and the entire mature miR-193b sequence was deleted. Mice homozygous for the mutant miR-193b-365-1 locus were born at a Mendelian ratio and were overtly normal.
Figure 1. Features of the murine miR-193b-365-1 locus and molecular analyses of BAT in its absence. (A) The miR-193b-365-1 locus, located on chromosome 16, is positioned downstream of the Mkl2 gene and spans approximately 25 kbp. The structure of primary transcripts containing miR-193b and miR-365b sequences was determined using RNA-seq reads from interscapular BAT (BAT-WT) of two-day old mice. At least eight putative exons were identified that form several splice forms. Two distinct sets of H3K4me3 marks, obtained from publicly available ChIP-seq data sets in liver, embryonic stem cell (ESC) and mammary cells (MG) (blue box) indicate the presence of two distinct promoter/regulatory regions. None of the sequenced reads obtained from interscapular BAT from miR-193b−/− mice (BAT-Null) were mapped to this locus confirming complete deletion of miR-193b (asterisk). (B) Two different high-throughput sequencing approaches, mRNA-seq (whole transcriptome shotgun sequencing), and miR-seq (microRNA sequencing), revealed differentially expressed genes and microRNAs in the absence of miR-193b compared with wild-type. Blue and red dots indicate up- and downregulated genes (or miRs), respectively. (C) Expression ratios of top 50 highly abundant miRNAs (in descending order from left to right) between BAT-Null and BAT-WT are shown as log2-fold-change value. Red dashed lines indicate 2-fold changes. (D) A total of 87 genes were defined as miR-193b targets by at least four different algorithms via miRecords.Citation30 Heatmap shows expression levels of the predicted miR-193b target genes. (E) qRT-PCR results from Ucp1 and Prdm16 (n = 3). Ucp1 in miR 193b-/- (CT 17.57 ± 0.35) and control BAT (16.34 ± 0.39); Prdm16 in miR 193b-/- (26.93 ± 0.02) and control BAT (26.71 ± 0.47).
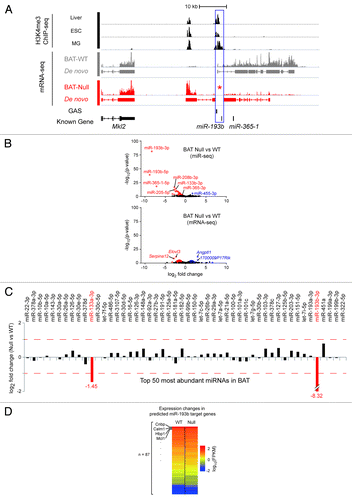
The absence of miR-193b and miR-365-1 in BAT from mutant mice was confirmed by miR-seq (; Table S1) and RNA-seq () analyses. MiR-Seq data also revealed that among the 50 most abundant miRs, only miR-133a was significantly deregulated in miR-193b-/- BAT (). RNA-seq of interscapular BAT of two-month old miR-193b-/- male mice verified the absence of transcripts over the deleted sequences (; Fig. S2). Transcription of the neighboring genes Mkl2 and Parn was not affected by the absence of miR-193b ( and GEO GSE47926).
Normal function of BAT in the absence of miR-193b
Visual inspection of two-month old miR-193b-/- male mice revealed the presence of interscapular BAT (n = 6) and no histological differences were observed between mutant and control tissues (). In addition, the respective weight of BAT, white fat, and different muscles was equivalent between control and miR-193b-/- male mice at the age of two months (Fig. S3). BAT function was determined by treating mice with the selective β3 adrenergic receptor agonist CL316.243 to activate BAT thermogenesis and by determining their cold tolerance. Total energy expenditure (TEE), as measured by indirect calorimetry, was comparable between control and mutant mice. Both groups doubled TEE in response to CL314.243, suggesting that the absence of miR-193b did not affect the thermogenic capacity of BAT (). To further determine whether BAT in miR-193b-/- mice was functional, their cold tolerance was assessed. Both control and experimental mice equally maintained their body temperature in a 4 °C environment over a period of 8 h ().
Figure 2. Brown adipose (BAT) development and function are unaffected by the absence of miR-193b/365-1. (A) H&E staining of BAT from two-month-old control (miR-193b+/+) and miR-193b/365-1 mutant (miR-193b-/-) males. (B) Effect of the β3 adrenergic agonist CL 316 243 on total energy expenditure in miR-193b+/+ and miR-193b-/- in two-month-old male mice (data are presented as mean +/− SEM (n = 6 mice/group). (C) Body temperature in two-month-old miR-193b+/+ and miR-193b-/- male mice exposed to 4°C for the indicated period of time. Values are mean ± SEM (n = 6). (D) Copy number of miR-193b and miR-193a per ng of total RNA isolated from BAT of two-month old miR-193b+/+ and miR-193b-/- mice. Values are mean ± SD (n = 3).
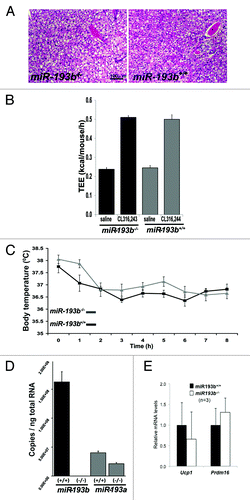
Since both miR-193a and miR-193b are highly expressed in BAT, the possibility remains that miR-193a could compensate for the absence of miR-193b. Although miR-193a is expressed at similar levels as miR-193b based on the miR-seq results (Table S1), when analyzed by qPCR, levels of miR-193a were 3-fold lower than miR-193b (5.0E+7 vs 1.5E+8) (). Overall, the miR-193a expression level was not elevated in miR-193b-/- BAT ( and ). The discrepancy between the miR-seq and the qPCR results might be attributed to the different ways in which microRNAs are detected in these two assays.Citation12
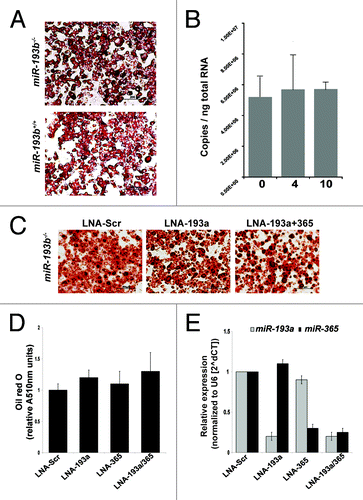
MiR-193a and miR-193b are encoded by two distinct genes and they possess an identical 8-nucleotide seed sequence. To determine whether the presence of miR-193a could compensate for the loss of miR-193b, we performed in vitro differentiation experiments using primary BAT cells. Oil red O staining determined that brown fat stromal vesicular fraction (SVF) cells isolated from miR-193b+/+ and miR-193b-/- newborn mice had the same capacity to differentiate in vitro (). In contrast to miR-193b,Citation6miR-193a levels did not increase in differentiating miR-193b-/- SVF cells (). When miR-193b-/- SVF cells were transfected with locked nucleic acids (LNAs) (scrambled, miR-193a and a combination of miR-193a and 365-1), individually or in combination, differentiation proceeded equally under all conditions (). The LNAs effectively inhibited the respective miRNAs as determined by qPCR (). Based on these experiments we conclude that miR-193a does not compensate for the absence of miR-193b.
Figure 3. Differentiation of miR-193b-/- pre-adipocytes is independent of miR-193a. (A) Oil-red-O staining of brown fat SVF cells, isolated from miR-193b+/+ and miR-193b-/- newborn mice, four days after differentiation. (B) Absolute quantification of miR-193a levels during adipogenesis (0, 4, and 10 d after induction) of brown fat SVF cells. Values are mean ± SD (n = 3). (C) SVF cells were transfected with LNA miRNA inhibitors (100 nM) one day before differentiation. Oil red O staining was used to determine lipid content after induction of adipogenesis. (D) Quantification of Oil-red-O staining in differentiated SVF cells (four days after differentiation). Values are mean ± SEM (n = 4). (E) MiRNA expression levels in BAT SVF cells 4 d after transfection with LNA miRNA inhibitors (100 nM). Values are mean ± SD (n = 3).
Gene expression in miR-193b-/- BAT
RNA-seq analyses were used to assess the expression of genes known to be linked to BAT physiology and to identify genuine miR-193b target mRNAs. Steady-state mRNA levels of Adipoc, Cebpα, Fabp4, Pparγ, Pparα, Dio2, Ucp1, Prdm16, and Cidea were indistinguishable between miR-193b-/- and miR-193b+/+ BAT (). Moreover, expression of Runx1T1, a putative miR-193b target that inhibits brown fat adipogenesis was not altered in miR-193b-/- BAT. Real-time qPCR experiments validated RNA-seq data and expression of Ucp1 and Prdm16 was equivalent in miR-193b+/+ and miR-193b-/- BAT ().
Table 1. Expression of genes characteristic for adipogenesis, brown fat and the myogenic lineage
Since previous in vitro results inferred a switch of cell identity from BAT to the muscle lineage in the absence of miR-193bCitation6, the expression of muscle markers in miR-193b+/+ and miR-193b-/- BAT was analyzed. Although several muscle-specific genes, including Cmk, Myf5, Myf6, Myod, and Myog were reported to be upregulated in vitro upon inactivation of miR-193b,Citation6 we failed to detect any increased expression of these genes in the in vivo setting (). Expression of Myf5, Myod1, and Myog was less than 1 FPKM, and thus, can be considered background. Similarly, real-time qPCR assays demonstrated that levels of these three mRNAs were at background.
RNA-seq data also revealed that among the ~22 500 annotated genes, expression of 43 genes was significantly altered in the absence of miR-193b (; Table S2). However, none of them matched the predicted miR-193b target genes. Additionally, none of the predicted miR-193b target genes showed significant changes in expression ().
Discussion
This in vivo study demonstrates that miR-193b and miR-365-1 are not required for the development, differentiation, and function of BAT in the mouse. Although the reasons for the discrepancy between this in vivo study and published in vitro experimentsCitation6 remain elusive, the fundamental difference between these two investigations lies in the approach used to manipulate miR-193b and miR-365-1 levels. While we inactivated the miR-193b-365-1 locus from the mouse genome, and thus, created a true miR-193b-null allele, which also fails to produce miR-365-1, the in vitro study was based on LNA miRNA inhibitors to reduce miR-193b and miR-365-1 levels in primary cells. Not only have miR-193b-/- mice functional BAT, brown SFV cells from these mice also differentiate into BAT in vitro. It could be argued that loss of miR-193b results in the compensatory increase of other miRNAs. However, expression of only one miRNA (miR-455) was increased in miR-193b-/- BAT and there is no evidence that it controls BAT physiology. Moreover, we did not see a shift of brown preadipocytes to cells with myogenic features in the absence of miR-193b as had been proposed.Citation6 We also did not observe increased expression of myogenic markers, such as MYF5 and MYOD1. Moreover, expression of the myogenic gene Myh1Citation13 and expression of several miRNAs associated with muscle development, such as miR-489,Citation14 miR-133b and miR-206Citation15, and miR-499Citation16 were reduced in miR-193b-/- BAT.
Although it is not uncommon for in vitro and in vivo experimental approaches to yield different results, the underlying molecular basis remains elusive. It is possible that miRNA-inhibiting tools used in in vitro studies, such as miRNA antagomirs or LNAs, might evoke ill-defined and non-physiological responses.Citation17 LNA effects can also depend on the dose and time of analysis post-transfection.Citation18 Of note, divergent results were obtained from experiments with miR-21 and miR-143/144 antagomirs and the respective mutant mice.Citation3 Similarly, miR-133a was shown to control myoblast proliferation in vitroCitation19 but in vivo skeletal muscle development and function appear to be unaffected in miR-133a transgenic mice.Citation20 It has been argued that the absence of defined physiological changes in gene-knockout mice could be due to compensatory changes in entire gene regulatory networks over the course of development.
Of note, miR-133a levels were reduced in BAT in the absence of miR-193b, suggesting a potential compensatory mechanism. MiR-133a expression decreases upon commitment and differentiation of BAT and has been linked to the concomitant induction of Prdm16.Citation10 Inhibition of miR-133a in vitroCitation7,Citation21 or in vivoCitation9 in gene-knockout mice promotes the differentiation of BAT from respective progenitor cells, including satellite cells.Citation21 The possibility that reduced levels of miR-133a could compensate for a loss of miR-193b in BAT differentiation should not be excluded and could be tested using transgenic expression of miR-133a in miR-193b-/- mice. However, browning of white fat is only observed if at least three of the four mir-133a1/2 alleles were deleted in vivo.Citation9 Moreover, based on miR-seq data, expression of miR-133a is more than 5000-fold lower than mir-193b making it less likely that its reduction could induce compensatory changes. Although our study, again, epitomizes the need to complement in vitro studies performed in cultured cells with genetic loss-of-function studies in the mouse, it becomes evident that the execution of biological programs in vitro and in vivo could be subject to different regulatory loops.
Materials and Methods
Generation of miR-193b mutant mice
Animals were handled and housed in accordance with guidelines of the NIH and all experiments were approved by the Animal Care and Use Committee of NIDDK. The targeting strategy was devised by Y Feuermann and L Hennighausen in consultation with Ingenious (Ingenious Targeting). Targeted ES cells and mice were generated by Ingenious. A BAC containing the miR-193b locus was isolated from a C57BL/6 library (BAC clone RP23: 239H18) (Fig. S1). An 11.570 kbp region used to construct the targeting vector was sub-cloned from this BAC clone. The region was designed such that the long homology arm extends 5.54 kbp 5′ to miR-193b sequences. The 2.83 kbp short homology arm ends 3′ to miR-193b. The LoxP/FRT flanked Neo cassette was inserted 603 bp downstream of miR-193b. The single loxP site, containing engineered KpnI, EcoRV, EcoRI, PstI, and SpeI sites for southern blot analysis, was inserted 179 bp 5′ of miR-193b. The targeted region flanked by loxP sites has a size of 3.19 kbp and includes miR-193b (ch16:10447114-10450305). Targeted iTL BA1 (C57BL/6N x 129/SvEv) hybrid embryonic stem cells were microinjected into C57BL/6 blastocysts. Resulting chimeras with a high percentage agouti coat color were mated to C57BL/6 mice expressing FLP recombinase to remove the Neo cassette. PCR was performed to detect the presence of the distal LoxP site using the following primers: 5′- AGAAGAGTGA TGGCATCTCT GACG -3′ and 5′- TCCGCCCATT CCTTGTCTGT CTTA -3′. This reaction amplifies a wild-type product 397 bp in size. The presence of a second PCR product 68 bp greater than the wild-type product indicates a positive loxP PCR. Conversion of the floxed allele into a null mutation was achieved by transmitting the floxed allele through the female germline of transgenic mice expressing an MMTV-Cre transgene (line A).Citation22 Female MMTV-Cre miR-193bf/+ mice were mated with miR-193bf/+ males to produce miR193b+/− offspring to segregate out the MMTV-Cre transgene and to produce miR193b-/- mice in the subsequent generation. The null allele was detected using the following primers: 5′- AGAAGAGTGA TGGCATCTCT GACG -3′ and 5′- GGGCTCATTG TGTCGCGTAA GG -3′. This reaction amplifies a wild-type product 487 bp in size.
Indirect calorimetry
Energy expenditure was measured in 2-month old males at 30 °C by indirect calorimetry (CLAMS, Columbus Instruments) with food and water provided ad libitum. After a 2-day adaptation, each mouse was injected with selective beta3 adrenergic agonist CL316.243 (Sigma, 1 mg/kg, i.p.) at 10 AM and saline on the following day. Data were collected for 3 h, starting 1 h post-injection, and are presented as mean +/− SEM (n = 6 mice/group).
Cold tolerance
To determine the effects of cold exposure, body temperature was measured using an animal rectal probe thermometer. Basal temperature was determined. Then animals were placed at 4 °C and body temperature was measured every hour up to 8 h.
Cell culture
Brown fat stromal vascular fraction (SVF) cells were isolated and cultured according to published methods with a few modifications.Citation23 Briefly, intrascapular BAT of newborn mice was collected and digested with collagenase A (2 mg/ml). SVF cells were collected by centrifugation and red blood cells lysed with ACK Lysing Buffer. Digested tissue was filtered through a 100 µm cell strainer. SVF cells were cultured in DMEM with 10% fetal bovine serum (FBS) (Invitrogen). Upon confluence, cells were induced to differentiate for 2 d with DMEM containing 10% FBS (Gemini Bio-Products), 850 nM insulin, 0.5 µM dexamethasone, 250 µM 3-isobutyl-1-methylxanthine, phosphodiesterase inhibitor (IBMX), 1 µM Rosiglitazone, 1 nM T3 and 125 nM indomethacin. Subsequently, the medium was replaced with DMEM containing 10% FBS and 160 nM insulin for 2 d. To analyze lipid accumulation Oil red O staining was performed on day 4. Cells were fixed for 15 min with 10% formalin, washed with water, stained for 1 h in Oil red O stain and rinsed with water. Oil red O was quantified according to published methods.Citation24 In brief, Oil red O was extracted from the cells using iso-propanol and the absorbance was measured at 510 nm. Images were taken from all three experiments and representative examples are shown in .
FACS sorting
Primary brown fat cells were isolated as previously described.Citation6 For the staining, we used FITC-conjugated anti-CD45 (clone30-F11, eBioscience), APC-conjugated anti-CD11b (clone M1/70, eBioscience), and PE-conjugated anti-Ly-6A/E (Sca1) (clone D7, eBioscience) antibodies. Cells were washed and resuspended in FACS buffer (10% FCS, 1mM EDTA, 0.1% sodium azide). CD45− CD11b− Sca1+ preadipocytes were sorted with an ARIA II FACS instrument (BD Bioscience) in growth medium using a 100 µm nozzle and a pressure of 20 psi. For sorting and data analysis DIVA software (version 6.1.3, BD Bioscoience) was used.
Transfection of LNA miRNA inhibitors
Sorted brown fat SVF cells were grown to 80% confluence in DMEM with 10% FBS, LNA miRNA inhibitors (100 nM) were transfected by Lipofectamine® 2000 Transfection Reagent (Life Technologies: 11668-019) according to the manufacturer's instruction. Twenty-four h after transfection, cells were recovered in full culture medium and grown to confluence for differentiation. LNA inhibitors were purchased from Exiqon: LNA-365 (410222-04), LNA-193a (412847-04), and LNA-Ctl (199004-04).
Quantitative rtPCR assays
Real-time qPCR assays were performed for miR193a, miR193b, and miR365, and Ucp1 and Prdm16. MiRNA was isolated using the miRNeasy kit (Qiagen). Ten ng total RNA was used for the RT reaction using TaqMan MicroRNA RT Kit (ABI) according to the manufacturer’s instructions. The following TaqMan MicroRNA Assays (ABI) were used for the specific reverse transcription of mature miRNAs: mmu-miR-193b-3p Assay ID:002467, hsa-miR-193a-3p Assay ID: 002250, hsa-miR-365 Assay ID: 001020, snoRNA202 Assay ID: 001232. QPCR was performed with TaqMan® Universal Master Mix II, no UNG (ABI) according to the manufacturer’s protocol on an ABI PRISM 7900HT Sequence Detection System. Data were analyzed using the SDS software (Ver. 2.3). For absolute quantification of miRNAs, synthetic single-stranded RNA oligonucleotides corresponding to the mature miRNA sequence (miRBase Release 19) were used (Invitrogen). SnoRNA202 was used for normalization.
Real-time RT-PCR was performed to validate expression of Ucp1 and Prdm16.
Small RNA-seq
Total RNA was extracted using the miRNeasy Mini Kit (Qiagen), Small RNA libraries were prepared with a small RNA version 1.5 sample preparation kit (Illumina). Total RNA was ligated with adaptors specifically modified to target microRNAs (supplied with the sample preparation kit). Reverse transcription (RT)-PCR amplification was performed using the adaptors as primers. The resulting double-stranded cDNA libraries were purified by PAGE using 6% Novex Tris-borate-EDTA gels (Invitrogen) and size selected to eliminate dimerized adaptors. The quality and concentration of libraries were determined by an Agilent 2100 Bioanalyzer and RiboGreen fluorescence on QuBit (Invitrogen). The libraries were sequenced using HiSeq 2000 (Illumina). To estimate relative expression levels of miRs in each sample, the number of sequenced single-end reads overlapped with annotated miRs was estimated by miRDeep2.Citation25 Relative expression levels of miRs were calculated by means of RPM (reads per million). Extremely low abundant miRs in both BAT-WT and BAT-Null (mean RPM below 5) were discarded. Differentially expressed miRs were identified by DESeq with an adjusted p-value cutoff of 0.05 (Table S1). The miRNA-seq data are deposited in GEO under accession number (GSE47926).
RNA-seq
Total RNA was extracted using the miRNeasy Mini Kit (Qiagen), Poly(A) RNA was purified from 1 µg total RNA and cDNA was synthesized using SuperScript II (Invitrogen) and TruSeq RNA Sample Preparation Kit (Illumina Inc.), and sequenced using HiSeq 2000 (Illumina). The single-end reads of each sample were aligned to the mouse reference genome (mm9 assembly) using the TopHat program.Citation26 Significantly up- and downregulated genes were independently identified by DESeq, edgeR, and Cufflinks with an adjusted p-value cutoff (< 0.05) and merged into one.Citation27-Citation29 Among the genes, we defined differentially expressed genes (DEGs) as the genes detected by at least two different algorithms (Table S2). To detect primary transcripts originated from the Mir-193b-365-1 locus, we reconstructed a transcriptome by using a de novo transcriptome assembly program, Cufflinks. The RNA-seq data are deposited in GEO under accession number (GSE47926).
Histology
Histological images from interscapular BAT were obtained from six control males and six miR-193b-/- males at 2 mo of age. shows representative images.
Additional material
Download Zip (609.2 KB)Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by the NIDDK and NCI Intramural Research Programs. We acknowledge Tatyana Chanturiya for technical support, Kai Ge (NIDDK) and Vittorio Sartorelli (NIAMS) for reagents and advice, Marc Reitman for scientific input and Michael Rieger for comments. N.H. was supported by the German Jose Carreras Leukemia Foundation (DJCLS R11/02) and by the LOEWE Center for Cell and Gene Therapy Frankfurt, Hessisches Ministerium für Wissenschaft und Kunst (III L 4- 518/17.004 (2010) and the Georg-Speyer-Haus.
Contributions
Y Feuermann designed and performed experiments (histology, RNA-seq) and analyzed data; K Kang performed computations analyses; O Gavrilova designed and performed the metabolic experiments; N Haetscher designed and performed miRNA real-time expression experiments; KH Yoo designed and performed real-time qPCR experiments; S Jin Jang performed cold tolerance tests and genotyped mice; C Jiang performed in vitro BAT differentiation experiments; FJ Gonzalez analyzed and interpreted data; GW Robinson analyzed and interpreted data; L Hennighausen designed experiments, analyzed, and interpreted data. This research has been performed by the NIH. All authors participated in writing and editing the manuscript.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/rnabiology/article/27239/
References
- Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature 2008; 455:64 - 71; http://dx.doi.org/10.1038/nature07242; PMID: 18668037
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010; 466:835 - 40; http://dx.doi.org/10.1038/nature09267; PMID: 20703300
- Patrick DM, Montgomery RL, Qi X, Obad S, Kauppinen S, Hill JA, van Rooij E, Olson EN. Stress-dependent cardiac remodeling occurs in the absence of microRNA-21 in mice. J Clin Invest 2010; 120:3912 - 6; http://dx.doi.org/10.1172/JCI43604; PMID: 20978354
- Morrisey EE. The magic and mystery of miR-21. J Clin Invest 2010; 120:3817 - 9; http://dx.doi.org/10.1172/JCI44596; PMID: 20978356
- Mudhasani R, Puri V, Hoover K, Czech MP, Imbalzano AN, Jones SN. Dicer is required for the formation of white but not brown adipose tissue. J Cell Physiol 2011; 226:1399 - 406; http://dx.doi.org/10.1002/jcp.22475; PMID: 20945399
- Sun L, Xie H, Mori MA, Alexander R, Yuan B, Hattangadi SM, Liu Q, Kahn CR, Lodish HF. Mir193b-365 is essential for brown fat differentiation. Nat Cell Biol 2011; 13:958 - 65; http://dx.doi.org/10.1038/ncb2286; PMID: 21743466
- Trajkovski M, Ahmed K, Esau CC, Stoffel M. MyomiR-133 regulates brown fat differentiation through Prdm16. Nat Cell Biol 2012; 14:1330 - 5; http://dx.doi.org/10.1038/ncb2612; PMID: 23143398
- Trajkovski M, Lodish H. MicroRNA networks regulate development of brown adipocytes. Trends Endocrinol Metab 2013; 24:442 - 50; http://dx.doi.org/10.1016/j.tem.2013.05.002; PMID: 23809233
- Liu W, Bi P, Shan T, Yang X, Yin H, Wang YX, Liu N, Rudnicki MA, Kuang S. miR-133a regulates adipocyte browning in vivo. PLoS Genet 2013; 9:e1003626; http://dx.doi.org/10.1371/journal.pgen.1003626; PMID: 23874225
- Seale P, Conroe HM, Estall J, Kajimura S, Frontini A, Ishibashi J, Cohen P, Cinti S, Spiegelman BM. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J Clin Invest 2011; 121:96 - 105; http://dx.doi.org/10.1172/JCI44271; PMID: 21123942
- Wagner KU, Wall RJ, St-Onge L, Gruss P, Wynshaw-Boris A, Garrett L, Li M, Furth PA, Hennighausen L. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res 1997; 25:4323 - 30; http://dx.doi.org/10.1093/nar/25.21.4323; PMID: 9336464
- Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet 2012; 13:358 - 69; http://dx.doi.org/10.1038/nrg3198; PMID: 22510765
- Wang M, Yu H, Kim YS, Bidwell CA, Kuang S. Myostatin facilitates slow and inhibits fast myosin heavy chain expression during myogenic differentiation. Biochem Biophys Res Commun 2012; 426:83 - 8; http://dx.doi.org/10.1016/j.bbrc.2012.08.040; PMID: 22910409
- Cheung TH, Quach NL, Charville GW, Liu L, Park L, Edalati A, Yoo B, Hoang P, Rando TA. Maintenance of muscle stem-cell quiescence by microRNA-489. Nature 2012; 482:524 - 8; http://dx.doi.org/10.1038/nature10834; PMID: 22358842
- Koutsoulidou A, Mastroyiannopoulos NP, Furling D, Uney JB, Phylactou LA. Expression of miR-1, miR-133a, miR-133b and miR-206 increases during development of human skeletal muscle. BMC Dev Biol 2011; 11:34; http://dx.doi.org/10.1186/1471-213X-11-34; PMID: 21645416
- Chen Y, Gelfond J, McManus LM, Shireman PK. Temporal microRNA expression during in vitro myogenic progenitor cell proliferation and differentiation: regulation of proliferation by miR-682. Physiol Genomics 2011; 43:621 - 30; http://dx.doi.org/10.1152/physiolgenomics.00136.2010; PMID: 20841498
- Khan AA, Betel D, Miller ML, Sander C, Leslie CS, Marks DS. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat Biotechnol 2009; 27:549 - 55; http://dx.doi.org/10.1038/nbt0709-671a; PMID: 19465925
- Elmén J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, Hedtjärn M, Hansen JB, Hansen HF, Straarup EM, et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res 2008; 36:1153 - 62; http://dx.doi.org/10.1093/nar/gkm1113; PMID: 18158304
- Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet 2006; 38:228 - 33; http://dx.doi.org/10.1038/ng1725; PMID: 16380711
- Deng Z, Chen JF, Wang DZ. Transgenic overexpression of miR-133a in skeletal muscle. BMC Musculoskelet Disord 2011; 12:115; http://dx.doi.org/10.1186/1471-2474-12-115; PMID: 21615921
- Yin H, Pasut A, Soleimani VD, Bentzinger CF, Antoun G, Thorn S, Seale P, Fernando P, van Ijcken W, Grosveld F, et al. MicroRNA-133 controls brown adipose determination in skeletal muscle satellite cells by targeting Prdm16. Cell Metab 2013; 17:210 - 24; http://dx.doi.org/10.1016/j.cmet.2013.01.004; PMID: 23395168
- Wagner KU, McAllister K, Ward T, Davis B, Wiseman R, Hennighausen L. Spatial and temporal expression of the Cre gene under the control of the MMTV-LTR in different lines of transgenic mice. Transgenic Res 2001; 10:545 - 53; http://dx.doi.org/10.1023/A:1013063514007; PMID: 11817542
- Schulz TJ, Huang TL, Tran TT, Zhang H, Townsend KL, Shadrach JL, Cerletti M, McDougall LE, Giorgadze N, Tchkonia T, et al. Identification of inducible brown adipocyte progenitors residing in skeletal muscle and white fat. Proc Natl Acad Sci U S A 2011; 108:143 - 8; http://dx.doi.org/10.1073/pnas.1010929108; PMID: 21173238
- Ramírez-Zacarías JL, Castro-Muñozledo F, Kuri-Harcuch W. Quantitation of adipose conversion and triglycerides by staining intracytoplasmic lipids with Oil red O. Histochemistry 1992; 97:493 - 7; http://dx.doi.org/10.1007/BF00316069; PMID: 1385366
- Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res 2012; 40:37 - 52; http://dx.doi.org/10.1093/nar/gkr688; PMID: 21911355
- Trapnell C, Salzberg SL. How to map billions of short reads onto genomes. Nat Biotechnol 2009; 27:455 - 7; http://dx.doi.org/10.1038/nbt0509-455; PMID: 19430453
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010; 11:R106; http://dx.doi.org/10.1186/gb-2010-11-10-r106; PMID: 20979621
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010; 26:139 - 40; http://dx.doi.org/10.1093/bioinformatics/btp616; PMID: 19910308
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 2013; 31:46 - 53; http://dx.doi.org/10.1038/nbt.2450; PMID: 23222703
- Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res 2009; 37:D105 - 10; http://dx.doi.org/10.1093/nar/gkn851; PMID: 18996891