Abstract
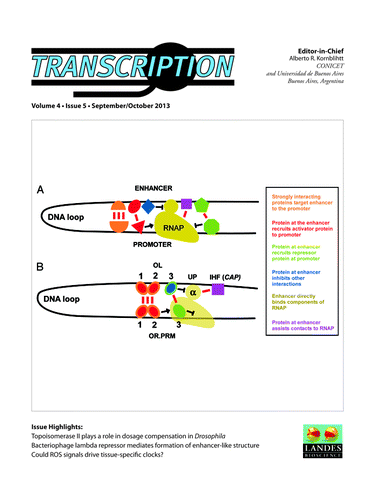
Unique role of SRSF2 in transcription activation
Target recognition of RNA-binding protein GLD-1
Ribosome-inactivating proteins
Large oncosomes mediate intercellular transfer of functional microRNA
Unique role of SRSF2 in transcription activation
Sudong Mo, Xiong Ji, and Xiang-Dong Fu
Transcription pause release from gene promoters has been recognized to be a critical point for transcriptional regulation in higher eukaryotes. Recent studies suggest that regulatory RNAs are extensively involved in transcriptional control, which may enlist various RNA binding proteins. Dr Xiang-Dong Fu and coworkers recently showed a key role of SRSF2, a member of the SR (for serine/arginine-rich) family of splicing regulators, in binding to promoter-associated small RNA to mediate transcription pause release, a regulatory strategy akin to the function of the HIV Tat protein via binding to the TAR element in nascent RNA to activate transcription. In a new study, the authors further dissect the structural requirement for SRSF2 to function as a transcription activator and extend the analysis to multiple SR proteins and hnRNPs (heterogeneous nuclear ribonucleoproteins). They showed that SRSF2 is a unique SR protein that activates transcription in a position-dependent manner while three other SR proteins enhance translation in a position-independent fashion. In contrast, multiple hnRNPs appeared to negatively influence mRNA levels, especially when tethered in the gene body. These findings suggest broad participation of RNA binding proteins in diverse aspects of regulated gene expression at both the transcriptional and posttranscriptional levels in mammalian cells.Citation1 ()
https://www.landesbioscience.com/journals/transcription/article/26932/
Target recognition of RNA-binding protein GLD-1
Jung H Doh, Yuchae Jung, Valerie J Reinke, and Min-Ho Lee
Germline and early development of various organisms is highly dependent on the temporal and spatial control of maternal gene products. In Caenorhabditis elegans, one of the major regulators of maternal mRNA expression in the germline is an RNA-binding protein, GLD-1 (GermLine Development defective). This maxi-KH motif containing RNA-binding protein has various functions mainly during female germ cell development, suggesting that it likely controls the expression of a selective group of maternal mRNAs. In order to gain insight into how GLD-1 specifically recognizes these mRNA targets, Dr Jung Doh and colleagues identified 38 biochemically proven GLD-1 binding regions from multiple mRNA targets that are among over 100 putative targets co-immunoprecipitated with GLD-1. Computational analyses revealed three over-represented and phylogenetically conserved sequence motifs. The authors found that two of the motifs, one of which is novel, are important for GLD-1 binding in four GLD-1 mRNAs. Furthermore, they demonstrated the ability of secondary structure to obscure a GLD-1 binding motif and that the functionality of a GLD-1 binding motif is dependent on the surrounding sequences. Taken together, the study data suggest that some mRNAs recruit GLD-1 by a distinct mechanism, which involves more than one sequence motif that needs to be embedded in the correct context and structural environment.Citation2 ()
https://www.landesbioscience.com/journals/worm/article/26548/
Ribosome-inactivating proteins
Matthew J Walsh, Jennifer E Dodd, and Guillaume M Hautbergue
Ribosome-inactivating proteins (RIPs) were first isolated over a century ago and have been shown to be catalytic toxins that irreversibly inactivate protein synthesis. Elucidation of atomic structures and molecular mechanism has revealed these proteins to be a diverse group subdivided into two classes. RIPs have been shown to exhibit RNA N-glycosidase activity and depurinate the 28S rRNA of the eukaryotic 60S ribosomal subunit. In a recent review, Dr Guillaume Hautbergue and colleagues compare archetypal RIP family members with other potent toxins that abolish protein synthesis: the fungal ribotoxins which directly cleave the 28S rRNA and the newly discovered Burkholderia lethal factor 1 (BLF1). BLF1 presents additional challenges to the current classification system since, like the ribotoxins, it does not possess RNA N-glycosidase activity but does irreversibly inactivate ribosomes. The authors further discuss whether the RIP classification should be broadened to include toxins achieving irreversible ribosome inactivation with similar turnovers to RIPs, but through different enzymatic mechanisms.Citation3 ()
https://www.landesbioscience.com/journals/virulence/article/26399/
Large oncosomes mediate intercellular transfer of functional microRNA
Matteo Morello, Valentina R Minciacchi, Paola de Candia, Julie Yang, Edwin Posadas, Hyung Kim, Duncan Griffiths, Neil Bhowmick, Leland WK Chung, Paolo Gandellini, Michael R Freeman, Francesca Demichelis, and Dolores Di Vizio
Prostate cancer cells release atypically large extracellular vesicles (EVs), termed large oncosomes, which may play a role in the tumor microenvironment by transporting bioactive molecules across tissue spaces and through the blood stream. In a new study, Dr Dolores Di Vizio and colleagues applied a novel method for selective isolation of large oncosomes applicable to human platelet-poor plasma, where the presence of caveolin-1-positive large oncosomes identified patients with metastatic disease. This procedure was also used to validate results of a miRNA array performed on heterogeneous populations of EVs isolated from tumorigenic RWPE-2 prostate cells and from isogenic non-tumorigenic RWPE-1 cells. The results showed that distinct classes of miRNAs are expressed at higher levels in EVs derived from the tumorigenic cells in comparison to their non-tumorigenic counterpart. Large oncosomes enhanced migration of cancer-associated fibroblasts (CAFs), an effect that was increased by miR-1227, a miRNA abundant in large oncosomes produced by RWPE-2 cells. The study findings suggest that large oncosomes in the circulation report metastatic disease in patients with prostate cancer, and that this class of EVs harbors functional molecules that may play a role in conditioning the tumor microenvironment.Citation4 ()
References
- Mo S, Ji X, Fu XD. Unique role of SRSF2 in transcription activation and diverse functions of the SR and hnRNP proteins in gene expression regulation. Transcription 2013; 4; In press http://dx.doi.org/10.4161/trns.26932
- Doh JH, Jung Y, Reinke VJ, Lee MH. C. elegans RNA-binding protein GLD-1 recognizes its multiple targets using sequence, context, and structural information to repress translation. Worm 2013; 2; In press http://dx.doi.org/10.4161/worm.26548
- Walsh MJ, Dodd JE, Hautbergue GM. Ribosome-inactivating proteins: Potent poisons and molecular tools. Virulence 2013; 4; In press http://dx.doi.org/10.4161/viru.26399; PMID: 24071927
- Morello M, Minciacchi VR, de Candia P, Yang J, Posadas E, Kim H, Griffiths D, Bhowmick N, Chung LWK, Gandellini P, et al. Large oncosomes mediate intercellular transfer of functional microRNA. Cell Cycle 2013; 12:3526 - 36; http://dx.doi.org/10.4161/cc.26539; PMID: 24091630