Abstract
Argonaute proteins (AGOs) are vital components of the RNA-induced silencing complex in gene silencing. AGOs are indispensable for microRNA (miRNA) biogenesis as well as function, and are intracellularly localized to both the cytoplasm and the nucleus. Cytoplasmic AGO-miRNA complexes are mainly involved in cleavage or translational repression of target mRNAs while the nuclear ones are engaged in transcriptional gene silencing, methylation, chromatin remodeling, and splicing. In insects, AGO1 and AGO2 are involved in RNA interference and miRNA pathways but the components involved in their trafficking between the nucleus and the cytoplasm are not known. In this study, we found that importin β-4 facilitates AGO1 distribution to the nucleus, which is regulated by aae-miR-981 miRNA. The results also revealed association of prohibitin with AGO1 that may play an important role in its stability. Importantly, we found that AGO1 distribution to the nucleus is blocked by Wolbachia, an endosymbiotic bacterium introduced into the Dengue vector, Aedes aegypti. Our results provide basic mechanisms on intracellular trafficking of AGO1 in insects and how this may be altered by Wolbachia, which may affect trafficking of miRNAs to the nucleus leading to alteration in epigenetic effects.
Keywords: :
Introduction
MicroRNAs (miRNA) are small non-coding RNAs of ~22 nucleotides that play significant roles in transcriptional or post-transcriptional gene silencing.Citation1,Citation2 Argonaute proteins (AGOs) are the core components of RNA-induced silencing complex (RISC), which are guided by the mature miRNA strand to the complementary target site in mRNA resulting in mRNA/protein down- or upregulation.Citation3,Citation4 The structural features of AGOs are characterized by PAZ, MID, and PIWI domains.Citation5,Citation6 The N-terminal PAZ domain binds to the 3′ hydroxylated end of the miRNA, and the MID domain anchors to the 5′ monophosphorylated end, while the rest of the interaction is based on the sugar-phosphate backbone of miRNA at a certain position that helps in binding to the target mRNA.Citation7 Several AGO proteins have been discovered from plants and animals with conserved sequences but functional diversification.Citation8,Citation9 In mammals, four AGOs (AGO1–4) have been well characterized that are involved in the miRNA pathway, and it was recently shown that miRNAs are randomly sorted to individual AGOs.Citation10 In Drosophila, there are five AGOs; AGO1–3, Piwi, and Aub, in which AGO1 and AGO2 are reported to be involved in miRNA and RNAi pathways, respectively.Citation11,Citation12 Only AGO2 in mammals and flies possesses slicer activity and can also be associated with the biosynthesis of miRNA bypassing the Dicer-1 processing step.Citation13-Citation15 In Neurospora crassa, only one AGO has been found that functions in the RNAi pathway.Citation16 In plants, 10 different AGOs (AGO1–10) have been characterized with AGO1 and AGO10 associated with miRNA-mediated gene regulation.Citation17,Citation18
In animals, canonical miRNA biosynthesis includes Drosha-mediated pre-miRNA synthesis from a primary transcript in the nucleus and pre-miRNA export to the cytoplasm where Dicer-1 cleaves it to miRNA/miRNA* duplex.Citation19 One or both strands of the duplex can be functional depending on their recruitment into AGO proteins. Mature miRNAs are present predominantly in the cytoplasm but many studies now have revealed that certain miRNAs can re-enter the nucleus.Citation20,Citation21 Accordingly, AGO proteins have been found to transport to the nucleus and shuttle miRNAs from the cytoplasm to the nucleus where they can regulate transcriptional gene silencing (TGS). In mammals, AGO1 and AGO2 are localized to the nucleus and in association with other proteins induce heterochromatinization resulting in TGS as well as alternative splicing.Citation22,Citation23 There are few reports on the mechanism of AGOs’ transport, which includes Exportin-5 serving as a pore and importin-8 in association with AGO2 facilitating its translocation into human cells.Citation24,Citation25
Wolbachia is an endosymbiotic gram-negative bacterium, which is estimated to infect over 40% of insect species.Citation26 It manipulates the host reproductive biology through a variety of strategies, including cytoplasmic incompatibility, male-killing, feminization, and parthenogenesis, and also provide direct mutualistic benefits to hosts in certain contexts.Citation27 Recently, a life-shortening strain of Wolbachia (wMelPop-CLA) was successfully introduced into the Dengue vector Ae. aegyptiCitation28 and was shown to inhibit replication of several arthropod-borne pathogens in the mosquito.Citation29Wolbachia infection has also been shown to lead to global changes in host gene expression as well as the host miRNA profile.Citation30-Citation32
Following our previous findings that Wolbachia manipulates Ae. aegypti’s miRNAs to facilitate its maintenance,Citation30,Citation33,Citation34 in this study, we investigated the distribution of AGO1 and AGO2 proteins in the cytoplasm and the nucleus in the mosquito cells in the presence (+Wol) or absence (-Wol) of Wolbachia. The two proteins have been shown to be important in miRNA biogenesis and function in insects.Citation12 We revealed that importin and prohibitin proteins are associated with AGO1 and facilitate its translocation to the nucleus. Furthermore, we discovered that AGO1 distribution is under the control of a miRNA, aae-miR-981, which is upregulated in +Wol cells. aae-miR-981, in turn, downregulates importin β-4, which results in the blockage of AGO1 translocation to the nucleus in +Wol cells.
Results
AGO1, but not AGO2, distribution to the nucleus is blocked in Wolbachia-infected cells
To investigate the effect of Wolbachia on distribution of AGO1 and AGO2 proteins within the cell, we studied their subcellular localizations in -Wol and +Wol (wMelPop-CLA) Aag2 cells derived from Ae. aegypti. Cytoplasmic and nuclear fractions of proteins from the two cell lines were extracted and analyzed on western blots using Drosophila melanogaster AGO1 and AGO2-specific antisera. In -Wol cells, AGO1 protein (98 kDa) was detected in both cytoplasmic and nuclear fractions, while it was only detectable in the cytoplasm of +Wol cells and not in the nucleus (), although the protein levels of AGO1 was the same in the cytoplasm from both cell lines when normalized against the cytoplasmic control protein GAPDH (). In the same extractions, AGO2 (110 kDa) was detected in the cytoplasmic and nuclear fractions of both -Wol and +Wol cell lines (). The same blot was probed with anti-GAPDH as well as anti-histone H3 antibodies to confirm that there was no mixing and equal loading of cytoplasmic and nuclear proteins, respectively. The AGO1 and AGO2 signals in the nucleus were always the same in replicate extractions and consistently in lower concentrations compared with the cytoplasmic concentration, hence the weaker signals. This is obvious when their nuclear signals are compared with that of histone probed on the same blot. The AGO1 result was also confirmed in whole Ae. aegypti mosquitoes with and without Wolbachia (). To find out if lack of AGO1 distribution to the nucleus in Wolbachia-infected mosquitoes is also seen in wMelPop-infected Drosophila flies, cytoplasmic and nuclear fractions were analyzed from flies. Western blot analysis showed the same pattern in Drosophila (). The results suggested that in the presence of Wolbachia, AGO1 distribution to the nucleus is specifically blocked.
Figure 1. AGO1 trafficking to the nucleus is blocked in +Wol Aag2 cells, Ae. aegypti, and Drosophila. (A) Western blot analysis for detection of AGO1 and AGO2 in cytoplasmic and nuclear fractions of -Wol and +Wol Aag2 cells using specific antisera to the proteins. Anti-GAPDH and anti-Histone H3 antibodies were used on the same blots to confirm that there was no mixing and equal loading of cytoplasmic and nuclear proteins, respectively. Graphs show AGO1 and AGO2 protein bands normalized to GAPDH and Histone H3 using imageJ software. (B) Western blot analysis of +Wol and -Wol female Ae. aegypti whole mosquitoes and D. melanogaster flies. The blots were probed with anti-AGO1 and anti-GAPDH antibodies. (C) Northern blot hybridization with three different miRNA probes in GFP, AGO1, and AGO2-silenced cells. The same blot was washed with 0.1% boiling SDS for stripping and reused for other probes. U6 shows equal loading.
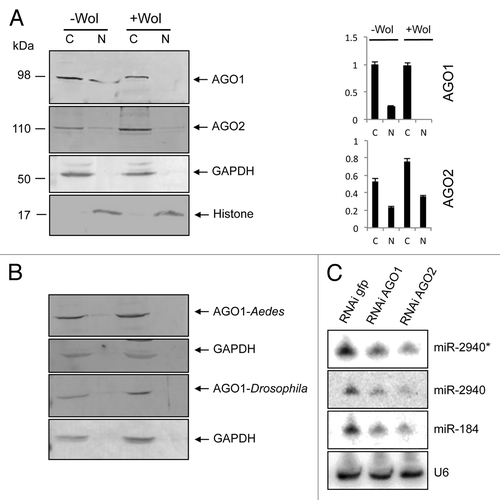
Interestingly, deep sequencing from cytoplasmic and nuclear RNA fractions of both -Wol and +Wol cells revealed the presence of some miRNAs such as aae-miR-2940* and -2940 in the cytoplasm and the nucleus of +Wol cells (data not shown). Based on our finding that AGO1 transport to the nucleus is reduced in +Wol cells, we hypothesized that these miRNAs found in the nucleus of +Wol cells were carried by AGO2. To test this, we used RNAi-mediated silencing of AGO1 and AGO2 genes since previous reports revealed that AGOs provide stability to mature miRNAs;Citation35 therefore, if AGO proteins are not available in the cell, miRNA levels must be reduced. Northern blot hybridizations showed substantial decline in aae-miR-2940*, -2940, and -184 levels in AGO2 silenced +Wol cells (). A decline in miRNA levels was also observed in AGO1-silenced cells. Together, these results suggest that miRNAs in +Wol cells can still be transported to the nucleus as they can be carried by AGO2 protein that was detected in the nucleus of both cell types.
Importin β-4 facilitates distribution of AGO1 to the nucleus
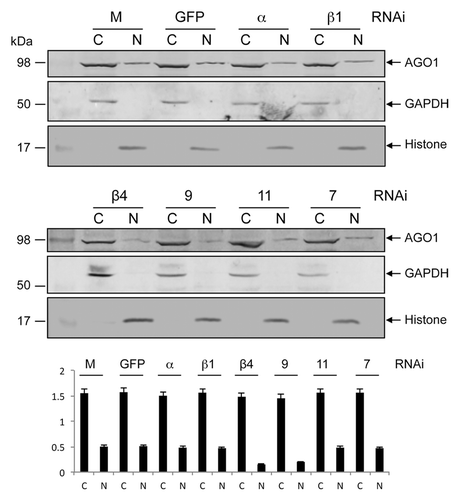
Based on previous reports on associated proteins involved in cellular distribution of AGO proteins,Citation25 we used RNAi-mediated gene silencing to study possible involvement of six Ae. aegypti importin proteins: importin α, β-1, β-4, 7, 9, and 11. We silenced all the six importin genes independently in -Wol Aag2 cells through in vitro synthesized sequence-specific dsRNAs, while using a non-homologous dsRNA for GFP as control. Gene silencing was confirmed through RT-qPCR using gene-specific primers, which showed significant reductions in the transcript levels of the silenced genes (Fig. S1). Western blot analyses showed that the overall level of AGO1 protein was the same in the cytoplasmic and nuclear fractions of importins α, β-1, 7, and 11 silenced cells when compared with control (). However, AGO1 protein level was notably reduced in the nuclear fraction of importins β-4- and 9-silenced cells, which suggested that their association with AGO1 might be important for its distribution to the nucleus. Since the effect of β-4 was slightly more pronounced and a monoclonal antibody was available to the protein, we selected this protein for further investigating its function.
Figure 2. RNAi-mediated silencing of importin genes to study their impact on AGO1 transport to the nucleus. Western blot analysis for the detection of AGO1 protein in cytoplasmic (C) and nuclear (N) fractions of -Wol cells transfected with mock, dsRNA of GFP, importin α, β-1 (top panel) β-4, 9, 11, and 7 (middle panel) using anti-AGO1 antibody. The same blots were probed with anti-GAPDH and anti-Histone H3 antibodies for cytoplasmic and nuclear protein levels. Lower panel graph shows normalization of AGO1 proteins to GAPDH and Histone H3 using imageJ software.
aae-miR-981 downregulates importin β-4
Our results from RNAi silencing of importin genes indicated that reduced amounts of importin β-4 protein led to reduced distribution of AGO1 in the nucleus of -Wol cells. We examined levels of importin β-4 protein using a monoclonal antibody to the protein in +Wol cells. Western blot analysis showed less importin β-4 protein in +Wol cells as compared with that in -Wol cells (), which is consistent with reduced AGO1 detection in the nucleus of +Wol cells. To find out if downregulation of importin β-4 is mediated by a cellular miRNA, we silenced Dicer-1 in +Wol cells using dsRNA specific to Ae. aegypti Dicer-1 sequence. Western blot analysis showed more importin β-4 protein in Dicer-1-silenced cells () supporting the likelihood that downregulation of importin β-4 is mediated by miRNA. Subsequently, RNAhybrid was used to find out potential targets in importin β-4 sequence against all Ae. aegypti miRNAs. In this search, we found a target sequence for aae-miR-981 with extensive complementarity and minimum free energy (mfe) of -22.6 Kcal/mol close to the stop codon in importin β-4 mRNA. To confirm this experimentally, we used synthetic aae-miR-981 mimic and a control mimic with random sequences for transfection into -Wol Aag2 cells. RT-qPCR analysis showed significant downregulation of importin β-4 transcript levels in aae-miR-981 mimic transfected cells as compared with that of control mimic (). In a separate experiment in -Wol cells, we confirmed that AGO1 distribution to the nucleus was reduced in aae-miR-981 mimic transfected cells replicating the observation in +Wol cells and mosquitoes (). These results confirmed that in +Wol cells AGO1 cellular translocation is under the control of aae-miR-981 via downregulation of importin β-4. We also confirmed upregulation of miR-981 in +Wol cells and mosquitoes through miRNA specific RT-qPCR ().
Figure 3. aae-miR-981 downregulates importin β-4. (A) Western blot analyses showing levels of importin β-4 protein using a specific monoclonal antiserum to the protein in cytoplasmic (C) and nuclear (N) fractions of -Wol and +Wol cells, and (B) in Dicer-1 silenced cells along with gfp control. (C) RT-qPCR analysis shows significant (*P < 0.05) downregulation of importin β-4 transcript in aae-miR-981 mimic-transfected cells (mimic) as compared with the control mimic (C mimic). Error bars represent standard errors of averages from three biological replicates. (D) AGO1 protein levels in cytoplasmic and nuclear fractions of -Wol cells in control mimic and aae-miR-981 mimic transfected cells. (B and C) were re-probed with anti-hsp70 antibody to show equal loading of the samples. (A and D) were re-probed with anti-GAPDH antibody to show equal loading of samples. Graphs indicate normalization of proteins to GAPDH and Histone H3 using imageJ software.
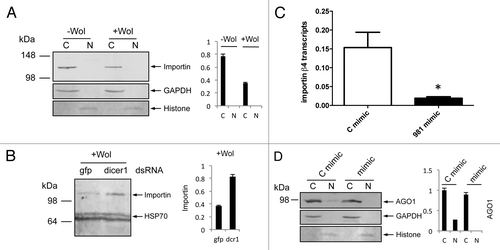
Figure 4. Target validation of aae-miR-981 in importin β-4. (A) miRNA-specific RT-qPCR analysis of RNA from -Wol and +Wol Aag2 cells and A. aegypti mosquitoes using a specific reverse oligo-dT primer and forward primer to aae-miR-981 normalized against 5S rRNA. Aae-miR-981 was significantly induced in the presence of Wolbachia (**P < 0.001). Error bars represent standard errors of averages from three biological replicates. (B) Cloning strategy of the target sequences complementary to the miRNA seed region from importin β-4 coding region in the 3′UTR region downstream of the GFP reporter gene ORF in the pIZ vector, denoted as pIZ/GFP-imp construct. The construct was co-transfected into -Wol cells along with aae-miR-981 mimic or the control mimic (C mimic). (C) RT-qPCR analysis indicated that GFP expression was significantly lower (***P < 0.0001) in aae-miR-981 mimic transfected cells as compared with control mimic (C mimic). Error bars represent standard errors of averages from three biological replicates.
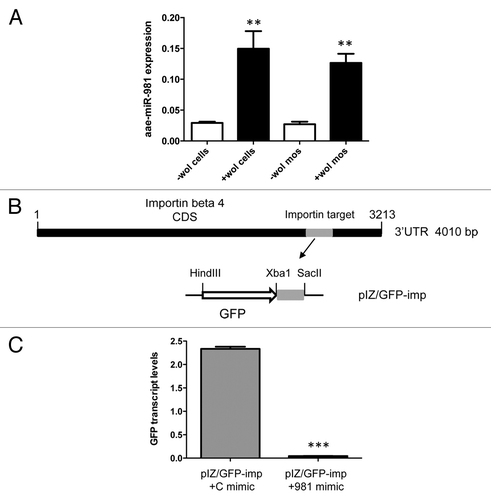
In order to further confirm the aae-miR-981-target interaction, we used a GFP-based reporter system in -Wol cells. First, the target sequence along with its flanking sequences from the coding region of importin β-4 (nucleotides 3072–3222) was cloned downstream of the GFP gene in the pIZ expression vector, resulting in the construct pIZ/GFP-imp (). The construct was co-transfected with aae-miR-981 and control mimics into -Wol cells. After 72 h, in aae-miR-981 mimic transfected cells, significantly lower GFP transcript levels were detected confirming downregulation of the target gene ().
Prohibitin associates with AGO1
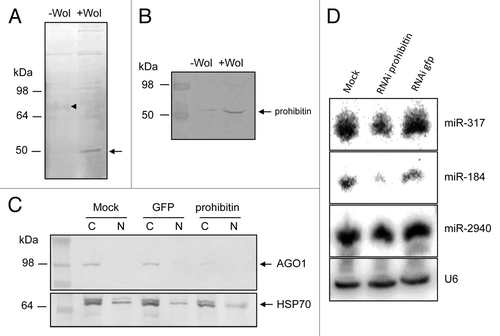
To find other proteins that could associate with AGO1 in -Wol and +Wol cells, we did immunoprecipitation (IP) of AGO1 using the specific Ae. aegypti AGO1 antiserum. After electrophoresis on a 10% protein gel and staining, a 50 kDa protein was found to be present in much higher quantities in +Wol cells as compared with -Wol cells and a fussy 72 kDa in reduced quantities in +Wol cells as compared with -Wol cells (). Although there were differences in the quantities of other proteins, the differences in the 50 and 72 kDa proteins was more pronounced and present in sufficient quantity for further analysis. Therefore, the proteins were selected and processed for peptide sequencing and identified as prohibitin (AAEL013952) for the 50 kDa and a number of hypothetical proteins for the 72 kDa protein through homology searches. Considering multi-functionality of prohibitin and its involvement in intracellular trafficking, subsequent experiments were focused on this protein. Western blot analysis using a prohibitin-specific antiserum showed a considerable upregulation of prohibitin in +Wol cells (), which was in agreement with the AGO1 IP gel (). Moreover, prohibitin was detected only in the cytoplasmic protein fractions in both cells (data not shown). We conducted RNAi-mediated silencing of prohibitin to see the impact on AGO1 transport in +Wol cells. Western blot analysis indicated significantly less AGO1 protein in prohibitin-silenced cells as compared with the controls (), which suggested that prohibitin might be important for the stability of AGO1. To confirm this, we did northern hybridization of small RNAs and found lower levels of aae-miR-317 and -184 but not -2940 in prohibitin-silenced cells (). This is consistent with our previous experiment where miR-2940 was suggested to be carried mainly by AGO2.
Figure 5. Prohibitin associates with AGO1. (A) Immunoprecipitation of AGO1 in -Wol and +Wol cells. A 50 kDa protein (arrow) was found in considerably higher quantities in +Wol cells and a 72 kDa (arrowhead) protein in much less quantities in +Wol cells. (B) Western blot analysis of -Wol and +Wol cells to detect prohibitin. (C) AGO1 protein levels in mock, GFP and prohibitin dsRNA-transfected +Wol cells; hsp70 shows equal loading. (D) Northern blot hybridization with three different miRNA probes in mock, prohibitin RNAi, and GFP RNAi in +Wol cells. The same blot was washed with 0.1% boiling SDS for stripping and reused for other probes. U6 shows equal loading.
Discussion
Recently, Argonautes (AGOs) have attracted considerable attention with their roles expanding in miRNA biogenesis, stability, and function as these proteins are at the heart of miRNA functionality. Several types of AGOs have now been characterized to associate with a variety of small RNAs from diverse organisms. A number of reports have revealed that in animal cells, AGOs are localized in the cytoplasm as well as in the nucleus, which opened up new avenues for detecting and investigating mature miRNAs inside the nucleus and their possible association with transcriptional gene silencing (TGS) and chromatin remodeling.Citation22 Here, we showed that in insects, AGO1 and AGO2 are normally found both in the cytoplasm and the nucleus, but Wolbachia blocks AGO1 transport to the nucleus in a cell line as well as mosquito and its natural host Drosophila. Previous investigations in Drosophila concluded sorting of miRNA and siRNA loading onto AGO1 and AGO2, respectively, whereas some guide miRNAs and many miRNA* were found loaded onto AGO2 as well.Citation12 High levels of some miRNAs, in the absence of AGO1 protein, in the nuclei of +Wol cells, suggested that these miRNAs might have been loaded onto AGO2. To confirm this, we did RNAi silencing of AGO1 and AGO2 independently and found lower levels of aae-miR-2940* and -2940 as compared with GFP dsRNA-transfected control cells. AGOs provide stability to mature miRNA and protect them from exonucleolytic degradation; therefore silencing of both proteins leads to less levels of miRNAs.Citation35 A recent report also highlighted quantitative functions of AGO proteins in mammalian cells where AGO2 is able to load more numbers of miRNAs because this protein is more abundant compared with AGO1 or AGO3.Citation10
Previous studies at the transcriptional level have shown that Wolbachia can have a significant impact on host gene expression.Citation31,Citation36 In order to find out the reason behind the lack of distribution of AGO1 to the nucleus in Wolbachia-infected cells, we investigated the role of a number of proteins that may facilitate AGO1 transport in mosquito cells, as this has not been investigated in any insects. On the basis of a recent finding that importin-8 was involved in the translocation of AGO2 from the cytoplasm to the nucleus in vertebrate cells,Citation25 we did RNAi-mediated silencing of six importin proteins from Ae. aegypti: α, β-1, β-4, 7, 9, and 11 in -Wol cells. Western blot analysis using AGO1-specific antisera showed less signal in the nuclear fraction from importins β-4- and 9-silenced cells, which suggested that these two importins are the most likely candidates to facilitate AGO1 trafficking. Consistent with this result, when specific antisera against importin β-4 were used as probe less importin β-4 was found in +Wol cells as compared with -Wol cells. Finding low levels of importin β-4 in +Wol cells indicated that the protein was downregulated in the presence of Wolbachia. Previously, we found that Wolbachia alters the host miRNA expression levels that regulate host genes.Citation30,Citation33 To find out if importin β-4 could possibly be downregulated by host miRNA(s), we first silenced Dicer-1 in +Wol cells and found higher amounts of importin β-4 in these cells as compared with control cells indicating involvement of host miRNAs in regulating the protein. Using bioinformatics, a potential target sequence was found in the coding region close to the stop codon in importin β-4 mRNA against aae-miR-981. Interestingly, this miRNA was found to be upregulated in +Wol mosquitoes relative to -Wol mosquitoes confirmed by miRNA-specific RT-qPCR analysis. We confirmed downregulation of importin β-4 by using aae-miR-981 synthetic mimic transfection into mosquito cells followed by RT-qPCR. In addition, using a GFP-based reporter construct in which the target sequences were cloned downstream to GFP, we confirmed specific interaction of aae-miR-981 with the target. In addition, AGO1 distribution to the nucleus was reduced in -Wol cells transfected with aae-miR-981, which verified the finding that importin β-4 is important for AGO1 translocation to the nucleus.
In order to explore other proteins associated with AGO1, we found a 50 kDa prohibitin protein enriched in immunoprecipitation (IP) of +Wol cells using AGO1-specific antiserum. Western blot analysis using the prohibitin antiserum confirmed upregulation of the protein in +Wol cells. Interestingly, silencing of prohibitin in +Wol cells led to a decrease in AGO1 protein levels as well as miRNA levels as shown by western blot analysis with anti-AGO1 antibody and northern blot with aae-miR-317 and -184, which are loaded onto AGO1. This indicated that prohibitin might provide structural or functional stability to AGO1. Prohibitin is a multifunctional protein known to be implicated in mitochondrial morphology and protein folding.Citation37 Chemical modifications during post-translational modification of AGO proteins alter their stability, subcellular localization, affinity to small RNAs, and their abundance.Citation38 For example, prolyl-4-hydroxylation at proline 700 protects AGOs from proteasome-mediated degradationCitation39 and phosphorylation of AGO2-conserved tyrosine 529 acts as a molecular switch that turns off AGO2’s affinity for small RNAs.Citation40
In conclusion, our results highlight a novel approach that Wolbachia utilizes to manipulate host gene expression by altering trafficking/distribution of AGO1 and miRNAs to the nucleus that can lead to differential gene expression, methylation, and chromatin remodeling of the host and possibly its own genome as well.
Materials and Methods
Insect cell lines
Ae. aegypti Aag2 cells were maintained in growth media in a 1:1 mixture of Mitsuhashi–Maramorosch and Schneider’s insect media (Invitrogen), supplemented with 10% FBS. Cells used for transfection and other experiments were at the log phase of proliferation. Ae. aegypti were reared at 25 °C with 80% relative humidity and a 12 h light regime. Aag2 cells infected with the wMelPop-CLA strain of Wolbachia (+Wol) were used as previously described.Citation30
Cytoplasmic and nuclear protein and RNA extractions
Cytoplasmic and nuclear fractions were isolated using PARISTM kit (Ambion) according to the manufacturer’s instructions. Briefly, cells were washed with PBS, pelleted at low speed, and gently resuspended in 300 µl of ice-cold cell fractionation buffer, incubated on ice for 10 min, centrifuged at 2000 rpm at 4 °C for 5 min. The pellet contained the nuclear fraction that was resuspended in 300 µl cell disruption buffer, vortexed vigorously, and was used as nuclear protein sample after adding 4 × SDS-PAGE loading buffer. The supernatant contained the cytoplasmic fraction that was mixed with 4 × SDS-PAGE buffer. RNA from both nuclear and cytoplasmic fractions was extracted with 2 × lysis/binding buffer followed by using filter cartridges and eluted in 50 µl heated elution buffer supplied in the kit.
RNAi-mediated silencing of cellular genes
Primer pairs both having T7 promoter sequences (5′-ATACGACTCA CTATAGGG-3′) were designed to amplify Ae. aegypti importins α (XM_001661088.1), β-1 (XM_001663385.1), β-4 (XM_001654693.1), 7 (XM_001659651.1), 9 (XM_001649090.1), 11 (XM_001648235.1), AGO1 (XM_001662504.1), AGO2 (FJ979880.1), and Dicer-1 (XM_001652162.1). For in vitro dsRNA synthesis, Megascript T7 kit (Ambion) was used. One μg of PCR product was incubated overnight at 37 °C, DNase-treated, and precipitated by the lithium chloride method following the manufacturer’s instructions (Ambion). A total of 4 μg of dsRNA was used in transfection of cells and cells were transfected again with the same reagents at 48 h after the first transfection. Cells were collected after 96 h for protein and RNA isolation. RT-qPCR was done to confirm transcript levels of silenced genes.
Immunoprecipitation (IP) and western blots
Ae. aegypti AGO1 IP was conducted according to the protocolCitation41 with few modifications. First, AGO1 antibody (3 µg) was added to 2 ml of RPMI medium and incubated with 60 μl of protein G Sepharose beads (GE Healthcare) and kept under constant rotation at 4 °C overnight. Second, cells were washed with 1 × PBS and lysed in 5 ml lysis buffer containing 25 mM TRIS-HCl (pH 7.5), 150 mM KCl, 2 mM EDTA, 0.5% NP-40, 0.5 mM DTT, and protease inhibitor Complete (Roche). Lysates were incubated for 30 min at 4 °C and cleared by centrifugation at 20 000 g for 30 min at 4 °C. Beads from overnight incubation were added to 5 ml of cell lysates for 2.5 h under constant rotation at 4 °C. After incubation, beads were washed twice with IP wash buffer (300 mM NaCl, 50 mM TRIS-HCl [pH 7.5], 5 mM MgCl2, 0.1% NP-40, 1 mM NaF) and subjected to 8% SDS-PAGE. For western blot detection, we used the primary antibodies anti-AGO1 and anti-AGO2 from Drosophila (Abcam), monoclonal anti-importin β-4 from Ae. aegypti (Abmart), anti-prohibitin (Santa Cruz Biotech), anti-HSP70 (Sigma), anti-GAPDH (Sigma), anti-Histone H3 (Invitrogen), and secondary alkaline phosphatase conjugated antibodies (rabbit, mouse) from Sigma. Specific protein bands were normalized against reference proteins using imageJ software (imagej.en.softonic.com).
Northern blot hybridizations
Total or fractionated RNA extracted from cells (30 μg) were run on 15% urea denaturing polyacrylamide gels, electroblotted to nylon membranes by a semi-dried western blotting apparatus (Bio-Rad), and UV cross-linked. DNA oligonucleotides (21–22 mer) reverse complementary to specific miRNA sequences were labeled with [α-32P]-dCTP using terminal nucleotide transferase. All probe hybridizations and washings were done under stringent conditions at 50 °C. Blots were exposed to a phosphorimager screen overnight, and radioactive signals were detected using a phosphorimager scanner (Storm, GE Healthcare). Blots were washed for removing old probe with boiling 0.1% SDS twice for 15 min each time. Stripping of the probe was confirmed by scanning the blots as described above.
miRNA target studies and RT-PCRs
RNAHybrid software was used to find potential targets of Ae. aegypti miRNAs in importin β-4 gene. The protein levels of target gene were assayed in western blot hybridization using a specific monoclonal antiserum. Target gene sequence of importin β-4 was PCR amplified with specific primers and cloned into pIZ/V5-His vector (Invitrogen) downstream to GFP using XbaI and SacII restriction sites resulting in pIZ/GFP-imp construct. Transcript levels of GFP were analyzed by RT-qPCR using gene-specific primers while utilizing the mosquito gene RPS17 as reference. For each experiment, two or three biological replicates with three technical replicates were analyzed in a Rotor-Gene thermal cycler (QIAGEN) under the following conditions: 95 °C for 30 s, and 40 cycles of 95 °C for 10 s, and 60 °C for 45 s, followed by the melting curve (68 °C to 95 °C). t test was used to compare differences in means between different treatments. aae-miR-981 mimic (UUCGUUGUCG ACGAAACCUG CA) and control scrambled mimic (UUCUCCGAAC GUGUCACGUT T) were synthesized by Genepharma and used in transfections in Aag2 cell line using the Cellfectin reagent (Invitrogen). Mimics are chemically synthesized double-stranded RNAs which mimic mature endogenous miRNAs after transfection into cells. RT-qPCR was performed to examine the levels of miR-981 by using a miRNA-specific oligo-dT primer for reverse transcription and a miRNA-specific sense as a forward primer as described previously, 5S rRNA was used for normalizing data in three biological replicates.
Additional material
Download Zip (131.7 KB)Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank David Morgenstern from the Institute of Molecular Bioscience at UQ for sequencing proteins. This project was supported by the National Health and Medical Research Council (APP1062983) to Asgari S and an Australian Research Council DECRA Fellowship (DE120101512) to Hussain M and a grant from the Foundation of the National Institutes of Health through the Grand Challenges in Global Health Initiative of the Bill and Melinda Gates Foundation to O’Neill SL.
References
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem 2010; 79:351 - 79; http://dx.doi.org/10.1146/annurev-biochem-060308-103103; PMID: 20533884
- Fagegaltier D, Bougé A-L, Berry B, Poisot E, Sismeiro O, Coppée J-Y, Théodore L, Voinnet O, Antoniewski C. The endogenous siRNA pathway is involved in heterochromatin formation in Drosophila.. Proc Natl Acad Sci U S A 2009; 106:21258 - 63; http://dx.doi.org/10.1073/pnas.0809208105; PMID: 19948966
- Asgari S. MicroRNA functions in insects. Insect Biochem Mol Biol 2013; 43:388 - 97; http://dx.doi.org/10.1016/j.ibmb.2012.10.005; PMID: 23103375
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009; 136:215 - 33; http://dx.doi.org/10.1016/j.cell.2009.01.002; PMID: 19167326
- Elkayam E, Kuhn C-D, Tocilj A, Haase AD, Greene EM, Hannon GJ, Joshua-Tor L. The structure of human argonaute-2 in complex with miR-20a. Cell 2012; 150:100 - 10; http://dx.doi.org/10.1016/j.cell.2012.05.017; PMID: 22682761
- Jinek M, Doudna JA. A three-dimensional view of the molecular machinery of RNA interference. Nature 2009; 457:405 - 12; http://dx.doi.org/10.1038/nature07755; PMID: 19158786
- Song J-J, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science 2004; 305:1434 - 7; http://dx.doi.org/10.1126/science.1102514; PMID: 15284453
- Höck J, Meister G. The Argonaute protein family. Genome Biol 2008; 9:210; http://dx.doi.org/10.1186/gb-2008-9-2-210; PMID: 18304383
- Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol 2008; 9:22 - 32; http://dx.doi.org/10.1038/nrm2321; PMID: 18073770
- Wang D, Zhang Z, O’Loughlin E, Lee T, Houel S, O’Carroll D, Tarakhovsky A, Ahn NG, Yi R. Quantitative functions of Argonaute proteins in mammalian development. Genes Dev 2012; 26:693 - 704; http://dx.doi.org/10.1101/gad.182758.111; PMID: 22474261
- Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell 2007; 130:299 - 308; http://dx.doi.org/10.1016/j.cell.2007.05.057; PMID: 17662944
- Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA 2010; 16:43 - 56; http://dx.doi.org/10.1261/rna.1972910; PMID: 19917635
- Förstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell 2007; 130:287 - 97; http://dx.doi.org/10.1016/j.cell.2007.05.056; PMID: 17662943
- Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science 2010; 328:1694 - 8; http://dx.doi.org/10.1126/science.1190809; PMID: 20448148
- Maurin T, Cazalla D, Yang S Jr., Bortolamiol-Becet D, Lai EC. RNase III-independent microRNA biogenesis in mammalian cells. RNA 2012; 18:2166 - 73; http://dx.doi.org/10.1261/rna.036194.112; PMID: 23097423
- Boland A, Huntzinger E, Schmidt S, Izaurralde E, Weichenrieder O. Crystal structure of the MID-PIWI lobe of a eukaryotic Argonaute protein. Proc Natl Acad Sci U S A 2011; 108:10466 - 71; http://dx.doi.org/10.1073/pnas.1103946108; PMID: 21646546
- Wang H, Zhang X, Liu J, Kiba T, Woo J, Ojo T, Hafner M, Tuschl T, Chua N-H, Wang X-J. Deep sequencing of small RNAs specifically associated with Arabidopsis AGO1 and AGO4 uncovers new AGO functions. Plant J 2011; 67:292 - 304; http://dx.doi.org/10.1111/j.1365-313X.2011.04594.x; PMID: 21457371
- Vaucheret H. Plant ARGONAUTES. Trends Plant Sci 2008; 13:350 - 8; http://dx.doi.org/10.1016/j.tplants.2008.04.007; PMID: 18508405
- Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol 2009; 11:228 - 34; http://dx.doi.org/10.1038/ncb0309-228; PMID: 19255566
- Jeffries CD, Fried HM, Perkins DO. Nuclear and cytoplasmic localization of neural stem cell microRNAs. RNA 2011; 17:675 - 86; http://dx.doi.org/10.1261/rna.2006511; PMID: 21363885
- Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science 2007; 315:97 - 100; http://dx.doi.org/10.1126/science.1136235; PMID: 17204650
- Ameyar-Zazoua M, Rachez C, Souidi M, Robin P, Fritsch L, Young R, Morozova N, Fenouil R, Descostes N, Andrau J-C, et al. Argonaute proteins couple chromatin silencing to alternative splicing. Nat Struct Mol Biol 2012; 19:998 - 1004; http://dx.doi.org/10.1038/nsmb.2373; PMID: 22961379
- Benhamed M, Herbig U, Ye T, Dejean A, Bischof O. Senescence is an endogenous trigger for microRNA-directed transcriptional gene silencing in human cells. Nat Cell Biol 2012; 14:266 - 75; http://dx.doi.org/10.1038/ncb2443; PMID: 22366686
- Castanotto D, Lingeman R, Riggs AD, Rossi JJ. CRM1 mediates nuclear-cytoplasmic shuttling of mature microRNAs. Proc Natl Acad Sci U S A 2009; 106:21655 - 9; http://dx.doi.org/10.1073/pnas.0912384106; PMID: 19955415
- Weinmann L, Höck J, Ivacevic T, Ohrt T, Mütze J, Schwille P, Kremmer E, Benes V, Urlaub H, Meister G. Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell 2009; 136:496 - 507; http://dx.doi.org/10.1016/j.cell.2008.12.023; PMID: 19167051
- Zug R, Hammerstein P. Still a host of hosts for Wolbachia: analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS One 2012; 7:e38544; http://dx.doi.org/10.1371/journal.pone.0038544; PMID: 22685581
- Werren JH, Baldo L, Clark ME. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol 2008; 6:741 - 51; http://dx.doi.org/10.1038/nrmicro1969; PMID: 18794912
- McMeniman CJ, Lane RV, Cass BN, Fong AW, Sidhu M, Wang YF, O’Neill SL. Stable introduction of a life-shortening Wolbachia infection into the mosquito Aedes aegypti.. Science 2009; 323:141 - 4; http://dx.doi.org/10.1126/science.1165326; PMID: 19119237
- Moreira LA, Iturbe-Ormaetxe I, Jeffery JA, Lu G, Pyke AT, Hedges LM, Rocha BC, Hall-Mendelin S, Day A, Riegler M, et al. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell 2009; 139:1268 - 78; http://dx.doi.org/10.1016/j.cell.2009.11.042; PMID: 20064373
- Hussain M, Frentiu FD, Moreira LA, O’Neill SL, Asgari S. Wolbachia uses host microRNAs to manipulate host gene expression and facilitate colonization of the dengue vector Aedes aegypti. Proc Natl Acad Sci U S A 2011; 108:9250 - 5; http://dx.doi.org/10.1073/pnas.1105469108; PMID: 21576469
- Kambris Z, Blagborough AM, Pinto SB, Blagrove MSC, Godfray HCJ, Sinden RE, Sinkins SP. Wolbachia stimulates immune gene expression and inhibits plasmodium development in Anopheles gambiae.. PLoS Pathog 2010; 6:e1001143; http://dx.doi.org/10.1371/journal.ppat.1001143; PMID: 20949079
- Kambris Z, Cook PE, Phuc HK, Sinkins SP. Immune activation by life-shortening Wolbachia and reduced filarial competence in mosquitoes. Science 2009; 326:134 - 6; http://dx.doi.org/10.1126/science.1177531; PMID: 19797660
- Zhang G, Hussain M, O’Neill SL, Asgari S. Wolbachia uses a host microRNA to regulate transcripts of a methyltransferase, contributing to dengue virus inhibition in Aedes aegypti.. Proc Natl Acad Sci U S A 2013; 110:10276 - 81; http://dx.doi.org/10.1073/pnas.1303603110; PMID: 23733960
- Osei-Amo S, Hussain M, O’Neill SL, Asgari S. Wolbachia-induced aae-miR-12 miRNA negatively regulates the expression of MCT1 and MCM6 genes in Wolbachia-infected mosquito cell line. PLoS One 2012; 7:e50049; http://dx.doi.org/10.1371/journal.pone.0050049; PMID: 23166816
- Winter J, Diederichs S. Argonaute proteins regulate microRNA stability: Increased microRNA abundance by Argonaute proteins is due to microRNA stabilization. RNA Biol 2011; 8:1149 - 57; http://dx.doi.org/10.4161/rna.8.6.17665; PMID: 21941127
- Rancès E, Ye YH, Woolfit M, McGraw EA, O’Neill SL. The relative importance of innate immune priming in Wolbachia-mediated dengue interference. PLoS Pathog 2012; 8:e1002548; http://dx.doi.org/10.1371/journal.ppat.1002548; PMID: 22383881
- Nijtmans LG, de Jong L, Artal Sanz M, Coates PJ, Berden JA, Back JW, Muijsers AO, van der Spek H, Grivell LA. Prohibitins act as a membrane-bound chaperone for the stabilization of mitochondrial proteins. EMBO J 2000; 19:2444 - 51; http://dx.doi.org/10.1093/emboj/19.11.2444; PMID: 10835343
- Giorgi C, Cogoni C, Catalanotto C. From transcription to translation: new insights in the structure and function of Argonaute protein. BMC Genomics 2012; 3:545 - 59
- Qi HH, Ongusaha PP, Myllyharju J, Cheng D, Pakkanen O, Shi Y, Lee SW, Peng J, Shi Y. Prolyl 4-hydroxylation regulates Argonaute 2 stability. Nature 2008; 455:421 - 4; http://dx.doi.org/10.1038/nature07186; PMID: 18690212
- Rüdel S, Wang Y, Lenobel R, Körner R, Hsiao HH, Urlaub H, Patel D, Meister G. Phosphorylation of human Argonaute proteins affects small RNA binding. Nucleic Acids Res 2011; 39:2330 - 43; http://dx.doi.org/10.1093/nar/gkq1032; PMID: 21071408
- Beitzinger M, Peters L, Zhu JY, Kremmer E, Meister G. Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biol 2007; 4:76 - 84; http://dx.doi.org/10.4161/rna.4.2.4640; PMID: 17637574