Abstract
Trypanosome gene expression is regulated almost exclusively at the posttranscriptional level, through mRNA stability, storage and degradation. Here, we characterize the ribonucleoprotein complex (mRNPs) corresponding to the zinc finger protein TcZC3H39 from T. cruzi comparing cells growing in normal conditions and under nutritional stress. The nutritional stress is a key step during T. cruzi differentiation from epimastigote form to human infective metacyclic trypomastigote form. The mechanisms by which the stress, altogether with other stimuli, triggers differentiation is not well understood. This work aims to characterize the TcZC3H39 protein during stress response. Using cells cultured in normal and stress conditions, we observed a dynamic change in TcZC3H39 granule distribution, which appeared broader in stressed epimastigotes. The protein core of the TcZC3H39-mRNP is composed of ribosomes, translation factors and RBPs. The TcZC3H39-mRNP could act sequestering highly expressed mRNAs and their associated ribosomes, potentially slowing translation in stress conditions. A shift were observed in the mRNAs associated with TcZC3H39: the number of targets in unstressed epimastigotes was smaller than that in stressed parasites, with no clear functional clustering in normal conditions. By contrast, in stressed parasites, the targets of TcZC3H39 were mRNAs encoding ribosomal proteins and a remarkable enrichment in mRNAs for the cytochrome c complex (COX), highly expressed mRNAs in the replicative form. This identification of a new component of RNA granules in T. cruzi, the TcZC3H39 protein, provides new insight into the mechanisms involved in parasite stress responses and the regulation of gene expression during T. cruzi differentiation.
Introduction
Trypanosoma cruzi, the causal agent of Chagas disease, has a complex life cycle, alternating between two hosts and with at least four defined developmental stages.Citation1 The shift between forms involves the expression of a specific set of genes at a particular point in the life cycle of the parasite.Citation2-Citation4 Gene expression in trypanosomatids differs from that in other eukaryotes, in that there is a lack of characteristic RNA pol II promoters, transcription is polycistronic and transcripts are processed by trans splicing.Citation5 The mRNAs of a given polycistronic unit may display different levels of expression, confirming that gene expression is regulated principally by posttranscriptional mechanisms.Citation5-Citation7 This regulation may occur at different levels, such as processing of the primary transcript, the transport of processed mRNAs from the nucleus to the cytoplasm, and modification of the distribution, stability and translation of mRNAs. The absence of transcriptional control make trypanosomes interesting models for studies of posttranscriptional regulation.
The mRNAs are always bound by RNA-binding proteins (RBPs), the combination of which determines the fate of the mRNA in the cell. These proteins are responsible for most of the events regulating mRNA fate in the cell. RBPs are widespread in diverse organisms; they constitute the seventh most abundant protein family in Homo sapiens, confirming their importance.Citation8,Citation9 It has been observed in various organisms that a subset of mRNAs encoding proteins with related functions are bound by specific RBPs to form ribonucleoprotein complexes (mRNPs); the mRNAs of a given mRNP are co regulated, determining whether they will be translated, stored or degraded, within the so-called “RNA regulon.”Citation10
In silico analysis has shown that trypanosomatids have similar numbers of CCCH-type zinc finger proteins to higher eukaryotes. However, only a few of these proteins have been characterized to date.Citation11 In T. cruzi, TcZFP1 has been shown to bind preferentially to C-rich regions,Citation12 whereas TcZFP2 has a higher affinity for A-rich regions and the mRNAs associated with this protein appear to be upregulated in metacyclic trypomastigotes, suggesting a possible role in parasite differentiation.Citation13 In T. brucei, the characterized zinc finger proteins are associated with parasite differentiation from the procyclic to bloodstream formsCitation14-Citation17 and the stability of their mRNA targets.Citation18-Citation20
Studies performed more than two decades ago provided evidence for the storage of functional mRNAs, not associated with polysomes, in the cytoplasm of T. cruzi.Citation21 In other eukaryotes, the different populations of mRNAsCitation22 are maintained in a dynamic equilibrium between translation (polysomal mRNA) and storage or commitment to degradation (post-polysomal mRNA).Citation10 We previously characterized more than 500 proteins bound to mRNA by LC/MS-MS on polysomal and polysome-free ribonucleoprotein complexes from T. cruzi epimastigotes and epimastigotes subjected to nutritional stress.Citation23 Many RBPs were identified, including TcZC3H39, a cytoplasmic CCCH zinc finger protein.
Here, we characterize the ribonucleoprotein complex associated with TcZC3H39 and its role in regulating gene expression in T. cruzi during stress response. Our results suggest that the TcZC3H39 protein is part of an mRNP targeting transcripts to downregulation, and that mRNAs associated with this mRNP encode proteins with related functions. This observation provides support for the existence of posttranscriptional regulons in T. cruzi, as a key element of the regulation of gene expression in this parasite.
Results
The TcZC3H39 protein
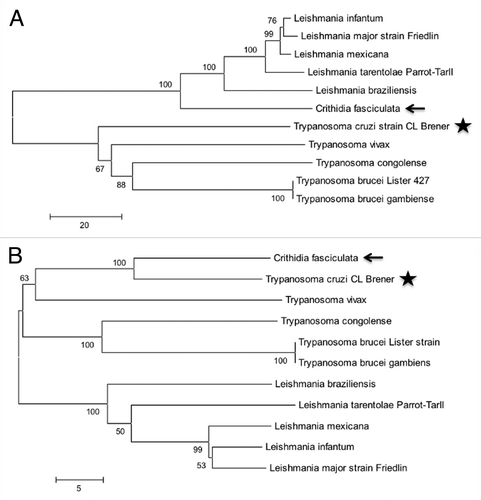
In an in silico study of the Trypanosoma brucei, T. cruzi and Leishmania major zinc finger proteins, the ZC3H39 protein was also named as CSBPA (cycling sequence binding protein A).Citation11 CSBPA was characterized in Crithidia fasciculata as a protein binding the 5′ untranslated (UTR) of specific transcripts regulated throughout the cell cycle and protecting these mRNAs from degradation before S phase.Citation24,Citation25 The CSBPA protein interacts with another protein with a very similar sequence, CSBPB. The genes encoding these two proteins are organized in tandem in the C. fasciculata genome and they display a high level of sequence identity, which led to the hypothesis that they resulted from gene duplication.Citation25 The CSBPA protein from C. fasciculata and the TcZC3H39 protein from T. cruzi have amino-acid sequences that are only 56% similar. However, a possible correlation of function was inferred for CSBPA and TcZC3H39 from a phylogenetic analysis on trypanosomatids ( and Fig. S1A). Analysis of the synteny downstream from the TcZC3H39 gene, identified TcZC3H40 as orthologous to Crithidia CSBPB ( and Fig. S1B). The CSBPA and TcZC3H39 proteins were found to have been present in the common ancestor of the Trypanosomatidae, but they diverged early as indicated by branch separation (). The domains of these proteins are conserved in all trypanosomatid orthologs studied to date, but the large number of substitutions per million years has resulted in a large evolutionary distance between these orthologs in analyses based on neighbor-joining methods (). This precluded inferences about the function of CSBPA in T. cruzi. In the case of CSBPB, the conservation of this second copy appears to have occurred only in C. fasciculata and T. cruzi. In other organisms, this protein has lost the U-box and CCCH domains; as a result, the degree of sequence identity is greatest between C. fasciculata and T. cruzi for this protein ( and Fig. S1C).
Figure 1. Phylogenetic analysis of TcZC3H39 . The trypanosomatid sequences were selected on the basis of BLASTp comparison results for TcZC3H39 (A) and TcZC3H40 (B). The proteins IDs are: LinJ.19.0290 (A) and LinJ.19.0280 (B) (Leishmania infantum), LmjF.19.0300 (A) and LmjF.19.0290 (B) (Leishmania major), LmxM.19.0300 (A) and LmxM.19.0290 (B) (Leishmania mexicana), LtaP.19.0260 LtaP.19.0250 (B) (Leishmania tarantolae), LbrM.19.0610 (A) and LbrM.19.0600 (B) (Leishmania brasiliensis), TvY486.1014340 (A) and TvY486.1014330 (B) (Trypanosoma vivax), TcIL3000.10.12810 (A) and TcIL3000.10.12800 (Trypanosoma congolense), Tbg972.10.18140 (A) and Tbg972.10.18130 (B) (Trypanosoma brucei gambiense), CfaC1_19_0320 (A) and CfaC1_19_0330 (B) (Crithidia fasciculata), Tb427.10.14930 (A) and Tb427.10.14290 (B) (Trypanosoma brucei) and TcCLB.506211.70 (A) and TcCLB.506211.60 (B) (Trypanosoma cruzi). Phylogenetic and molecular evolutionary analyses were conducted with MEGA version 5.Citation52 The sequences were aligned with ClustalW in the default configuration and constructed by the neighbor-joining method. The evolutionary distances indicated are the numbers of amino-acid substitutions per site. Bootstrap percentages (1000 replicas) are shown above the branches (if > 50). TcZC3H39 and TcZC3H40 are indicated by stars and CSBPA and CSBPB are indicated by arrows.
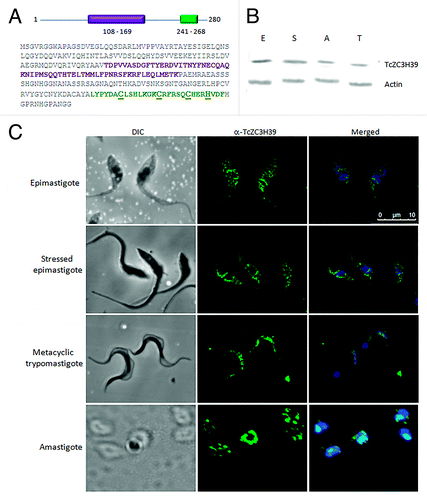
TcZC3H39 contains two domains: a U-box in the central portion and a CCCH-type domain in the C-terminal portion of the protein (). The U-box domain is one of the three domains of E3 ubiquitin protein ligases reported to be involved in substrate specificity for ubiquitination.Citation26 The presence of both ubiquitination domains and RNA-binding domains in the same protein has been described before.Citation27 Some proteins with CCCH domains, which are essential for oogenesis and
Figure 2. Expression pattern and cellular localization of TcZC3H39 during the life cycle of T. cruzi . (A) Diagram and amino-acid sequence of the domains of the TcZC3H39 protein. The U-box domain is shown in purple, the CCCH-type Zinc finger domain in green with the Cys and His underlined in bold. The numbers indicate the positions of the domains within the protein. (B) Western blot analysis of T. cruzi extracts. E: exponentially growing normal epimastigotes. S: epimastigotes subjected to nutritional stress. A: amastigotes. T: metacyclic trypomastigotes. The gels were normalized and 10 µg of protein were loaded into each lane. Top of the figure: anti-TcZC3H39 antibody (1:300); bottom of the figure, anti-actin was used as a normalizer (1:500), TcZC3H39 has an expected molecular mass of 31.1 kDa and actin, of 45.8 kDa. (C) Immunolocalization assay. Cells were incubated with anti-TcZC3H39 antibody (1:300). Immune complexes were detected by reaction with Alexa 488-labeled goat anti-mouse antibody (1:400). Kinetoplast and nuclei were stained with DAPI and the image obtained was merged with anti-TcZC3H39 images. Bar = 10 µm.
embryogenesis in C. elegans, are degraded in somatic cells by the ZIF-1 protein, through interactions between this protein and the CCCH domain, leading to the recruitment of an E3 ubiquitin ligase.Citation28
TcZC3H39 granule characterization
TcZC3H39 is constitutively produced throughout the lifecycle of the parasite, suggesting a role in all developmental stages of T. cruzi (). However the cellular localization pattern of TcZC3H39 is dynamic. A slightly granular pattern is observed in epimastigotes, which becomes more pronounced in stressed epimastigotes ( and Fig. S2). This response to stress conditions is similar to the mRNA mobilization occurring in RNA granules.Citation29-Citation31
Protein characterization of the TcZC3H39 granule
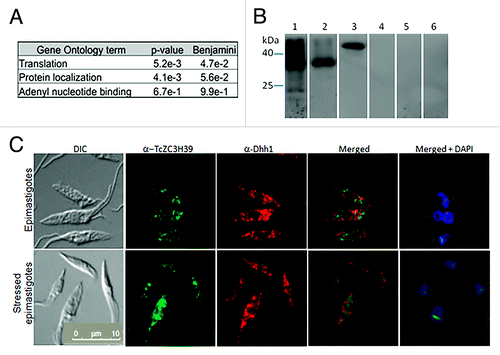
The proteins forming the complex containing TcZC3H39 were characterized by immunoprecipitation followed by mass spectrometry analysis. We minimized the risk of misidentification, by using three different controls: the pre-immune serum, the epimastigote total protein extract and a non related antibody, against TcOlla protein,Citation32 a GTP-binding protein that does not possess any RNA-binding domain. (Table S1 – raw data available upon request). An analysis of the proteins immunoprecipitated provided further insight into the complex function. The partners of TcZC3H39 were identified by LC-MS/MS in epimastigotes and stressed parasites. We identified 23 proteins associated with TcZC3H39, for which two of the controls used displayed enrichment. Interestingly, the RNA-binding protein TcCLB.506211.60, the TcZC3H40, partner of TcZC3H39 in C. fasciculata (CSBPB and CSBPA respectively) was identified. In this organism these proteins interact and the same might occur in T. cruzi, since TcZC3H39 and Tc3H40 are part of the same complex. Many of the proteins identified are associated with translation (e.g., 40S ribosomal proteins S23, S29 and S30, the Tu family of GTP-binding elongation factors and eukaryotic peptide chain release factor subunit 1) (Table S1). Proteins involved in mRNA metabolism were also identified, such as ATP-dependent RNA helicases and the RNA-binding proteins (Table S1). The functional annotation of the proteins associated to TcZC3H39 showed enrichment for translation, protein localization and adenyl nucleotide binding terms in gene ontology analysis (). No changes in protein composition were detected during the epimastigote stress response. One of the possible reasons for this is that we characterized only the core of the complex (data not shown). The selection criteria were very conservative, and it is therefore possible that some of the proteins associated with TcZC3H39 did not fulfill the stringency requirements and were excluded from the analysis. The results were validated by co-immunoprecipitation followed by western blotting with antibodies against the ribosomal proteins S7 and P0 and the control used was immunoprecipitation with the pre-immune serum (). The TcDhh1 protein, an ATP-dependent RNA helicase characteristic of P-bodies, was also used to validate the results () because it appeared to be more abundant in the TcZC3H39 immunoprecipitate than in pre-immune serum. However, it was excluded from Table S1 because the other control showed no enrichment for this protein. Nonetheless, immune co-localization assays also confirmed the interaction, with partial overlap both in epimastigotes and stressed epimastigotes (). To assess whether the TcZC3H39 complex formation would be due to mRNA or direct protein-protein interactions, the cell extract was treated with RNase A (Fig. S3). Some proteins remained associated to TcZC3H39, especially TcZC3H40, indicating direct protein-protein interaction. However, it was also observed that some proteins were not identified after the RNase treatment, indicating that some interactions in this complex are associated by mRNA binding thus forming ribonucleoprotein complexes – mRNPs (Table S1).
Figure 3. Proteins associated with TcZC3H39. (A) Gene Ontology classifications for the proteins associated with TcZC3H39, by biological process. (B) Co-immunoprecipitation assay with the TcZC3H39 protein in epimastigotes subjected to nutritional stress validated by western blotting. TcZC3H39 immunoprecipitation (IP) antibodies against TcZC3H39 (31.1 kDa, 1:300 dilution) and TcS7 (23.9 kDa, 1:400 dilution) (lane 1) and pre-immune serum immunoprecipitation as a control (lane 4). TcP0 (34.9 kDa, 1:400 dilution) antibody (lane 2) and the control (lane 5). TcDhh1 (46.7 kDa, 1:100 dilution) antibody (lane 3) and control (lane 6). Molecular weight is indicated in kDa. (C) Colocalization assays of TcZC3H39 with TcDhh1. The antiserum against TcZC3H39 was produced in mouse and tested at a dilution of 1:300. Antibodies against the TcDhh1 protein were produced in rabbit and used at a dilution of 1:100. Immune complexes were detected by reaction with Alexa 546-labeled goat anti-rabbit and Alexa 488-labeled goat anti-mouse antibodies (1:400). Kinetoplast and nuclei were stained with DAPI and the resulting image was merged with anti-Dhh1 anti-TcZC3H39 already merged images. Bar = 10 µm.
mRNAs associated with TcZC3H39
We also investigated the dynamics of mRNP formation during T. cruzi differentiation, by characterizing the mRNAs associated with TcZC3H39 in unstressed epimastigotes and epimastigotes subjected to nutritional stress (a key trigger of differentiation). We characterized the mRNAs associated with TcZC3H39 by immunoprecipitation assays performed in triplicate, followed by RNA-seq. As a control, extracts were incubated with the mouse pre-immune serum. The RNA-seq data is available at NCBI Sequence Read Archive database (SRA) under the accession number SRX255066. Mapping statistic details are provided in . In total, using a RPKM value greater than 100 and FDR ≤ 0.01, we identified 49 transcripts as more abundant, by a fold-change of at least four, in the TcZC3H39-mRNP in epimastigotes than in the control (Table S2). In analyses of stressed epimastigotes, 199 mRNAs were identified as being more abundant, by a factor of at least four, in the stressed cells TcZC3H39-mRNP than in the control (Table S3).
Table 1. Sequencing mapping statistics
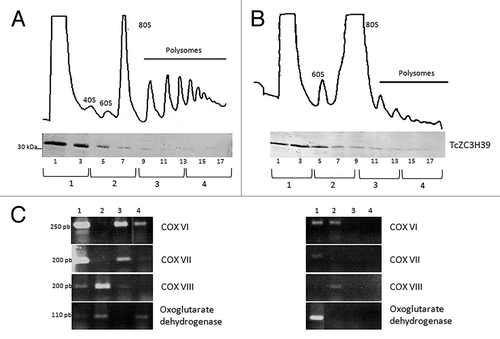
The results of ribonomics analyses were validated by immunoprecipitation followed by RT-PCR, with primers specific for the targets identified: dynein and MASP for unstressed epimastigotes and cytochrome c oxidase complexes VI, VIII, IX and oxoglutarate dehydrogenase for stressed parasites (). The controls for the RT-PCR assays were the RNAs immunoprecipitated with the pre-immune serum under the same conditions (). As another control to validate the data, we tested the mRNA targets of unstressed epimastigotes in stressed cell extracts and the stress-associated targets in unstressed epimastigote extracts (). The targets were detected by immunoprecipitation with TcZC3H39, but not by immunoprecipitation with the pre-immune serum. Thus, the targets associated with TcZC3H39 in unstressed epimastigotes were not associated with this protein in stress conditions and vice versa (). These results confirm that there is a shift in the population of mRNAs associated with TcZC3H39 in response to stress conditions.
Figure 4. Gene Ontology classification of unstressed and stressed epimastigotes transcripts associated with TcZC3H39 and RT-PCR validation of RNA-seq data. (A) Immunoprecipitation assays with exponentially growing normal epimastigotes (E), epimastigotes subjected to nutritional stress (S) and with the pre-immune serum and extracts of unstressed or stressed epimastigotes (PI E/S). Dynein, MASP, COX VI, COX VIII, COX IX and oxoglutarate dehydrogenase probes were used for RT-PCR validation. Size indicated in base pairs. (B) Gene Ontology classification of unstressed and stressed epimastigotes transcripts associated with TcZC3H39.
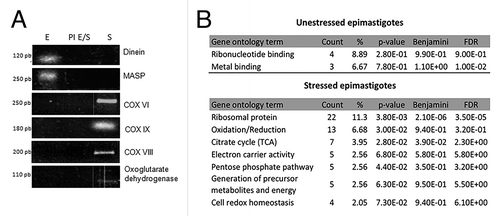
To study the possible functional coordination of the enriched transcripts, we investigated the Gene Ontology categories of the mRNAs associated with TcZC3H39 with the DAVID functional annotation tool. Grouping the mRNAs on the basis of Gene Ontology terms, we noticed a greater enrichment in specific terms for biological process in stressed epimastigotes than in unstressed epimastigotes. For unstressed epimastigotes, there was a slight enrichment in terms related to metal binding and ribonucleotide binding (). A major shift was observed in stressed epimastigotes, with a much higher proportion of mRNAs related to translation, the TCA cycle, cell redox homeostasis, electron carrier activity, oxidation/reduction, the pentose phosphate pathway, the generation of precursor metabolites and energy and ATPase activity (). Many cytochrome c oxidase (COX) subunits, including subunits V, VI, VIII and IX, as well as enrichment in transcripts encoding ribosomal proteins were identified by Kegg pathway analysis (figure not shown) These results confirm the correlation between the functions of the transcripts bound to TcZC3H39, supporting the RNA regulon theory, according to which, mRNAs encoding proteins with related functions are associated with specific proteins in mRNP complexes, which control their fate in the cell.Citation10,Citation33,Citation34
Downregulation of TcZC3H39 target mRNAs in stress
We investigated the destination of TcZC3H39 mRNA targets from the translation machinery in response to stress, by analyzing the association of transcripts with polysomes (). The mRNAs bound to TcZC3H39 in stress conditions were analyzed in epimastigotes in normal conditions () and under nutritional stress (). The sucrose gradient fractions were pooled separately into a light fraction (polysome-free), monosomes, light polysomes and heavy polysomes (). Under normal conditions, the COX VI, VII, VIII and oxoglutarate dehydrogenase transcripts were found to be associated with polysomes (). Proteomic analyses of parasites undergoing metacyclogenesis have identified these proteins in the epimastigote form, confirming that their mRNAs are actively translated at this developmental stage.Citation35 However, the analysis of these transcripts under conditions of nutritional stress indicated that they were present in the light or monosome fraction, indicating they were not translated and that their translation was apparently repressed by association with TcZC3H39 (). The protein is detected from the light to the heavier fractions of the gradient showing that it is part of large mRNP complexes that can go through the more concentrated sucrose fractions ().
Figure 5. Polysome fractionation (A) Unstressed epimastigotes. (B) Stressed epimastigotes. Upper figures represent the polysome profiles with each fraction indicated as follows: 1- Light fraction (Polysome-free); 2- Monosomes; 3- Light Polysomes; 4- Heavier Polysomes. Middle figures show the western blotting of each fraction using the anti-TcZC3H39 (1:300). The Molecular weight is indicated in KDa. (C) RT-PCR for each fraction, the size of the band is indicated for each gene.
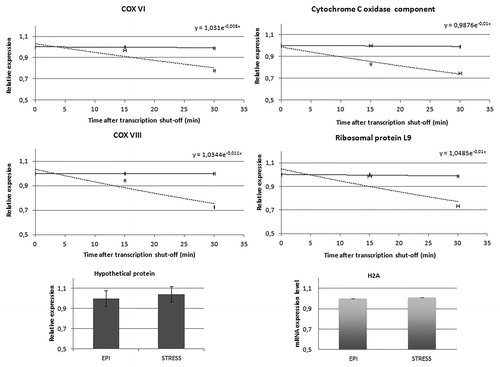
We then investigated the stability of the transcripts associated with TcZC3H39, using actinomycin D to inhibit transcription and comparing the decay of transcript levels for the putative COX VI, the COX VIII, the component protein of the cytochrome c oxidase complex and the L9 ribosomal protein in unstressed epimastigotes and stressed epimastigotes, by RT-qPCR (). Total cellular RNA was harvested 0, 15 and 30 min after the addition of actinomycin D and mRNA levels were measured by quantitative RT-PCR. mRNA decay half-lives were calculated based on a model of first order decay to be more than 86 min (82–90 min) for COX VI; 69 min (66–72 min) for the component protein of COX complex; 63 min (60 - 66 min) for the COX VIII and 69 min (67 - 71 min) for the L9 ribosomal protein mRNA in epimastigotes under nutritional stress. The difference in transcript half-lives between unstressed and stressed epimastigotes was statistically significant, with a P value of no more than 0.003, indicating that the mRNAs investigated were less stable in stressed than in unstressed epimastigotes. The decay analysis showed that the COX mRNAs were very stable in epimastigotes, consistent with their half-life of four hours reported for the procyclic form of T. brucei.Citation36 A comparison of unstressed and stressed epimastigotes showed transcript levels to be 20% lower in the stressed parasites, suggesting downregulation. To analyze if the downregulation was a general stress response for the transcripts in T. cruzi we used an mRNA not bound to TcZC3H39 as a control, which codes for a hypothetical protein and also the H2A transcript level, used for normalization of the targets. The results did not show any disturbance in the levels upon stress when compared with unstressed epimastigotes ().
Figure 6. Relative expression of TcZC3H39 target mRNAs. Total RNA was extracted from unstressed epimastigotes (continuous line) and stressed epimastigotes (dashed line) harvested 0, 15, and 30 min after the addition of actinomycin D. Reverse transcription-qPCR was performed according to MIQE criteria. Transcript levels at each time point were normalized to the corresponding level of the histone H2A transcript. The bar graphs in the lower panel refer to the expression levels of the hypothetical protein used as a control and the H2A mRNA levels in unstressed and stressed epimastigotes. Each point represents the mean and standard error of triplicate samples. A one-phase model of exponential decay was used to derive the decay curves for the indicated mRNAs.
Identification of elements in the 3′ UTR of the mRNAs bound to TcZC3H39
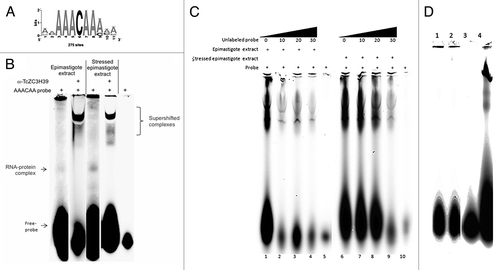
We searched for conserved regulatory sequences in the transcripts associated with TcZC3H39, by aligning 300 nucleotide sequences derived from the 3′ intergenic regions of the mRNAs after the CDS stop codon, using the motif discovery software of RSAT (Regulatory Sequence Analysis Tools - http://rsat.bigre.ulb.ac.be/).Citation37 The conserved hexamer motif AAACAA (e-value 2 e-13, ) was identified in the 3′ UTR region of most of the mRNAs (Table S4). There were between one and four repeats of this hexamer in the putative 3′UTR of the transcripts. A supershift assay was performed to check that TcZC3H39 recognized this motif. The TcZC3H39 protein bound this motif in vitro (), indicating that this element is one of the cis elements recognized by the protein for the modulation of target expression. The specificity of this recognition was confirmed in an assay including a nonspecific probe (). The A residues were replaced by U residues (UUUCUU). TcZC3H39 did not bind these probes, indicating that the A-rich region in the UTR of the transcript is, indeed, the binding site of the protein.
Figure 7. Supershift assay. (A) Consensus sequence obtained from the MEME and RSAT programs. (B) Supershift assay performed with the Cy5.5-labeled AAACAA probe, with enriched, unstressed and stressed epimastigote extracts. The black arrow indicates protein complexes without the antibody. (B) and (C) The supershifted complexes, RNA-protein complexes and the free probe are indicated in the figure. (D) Assay using a nonspecific probes changing A for U (UUUCUU). Lane 1 – Free probe (UUUCUU), lane 2– enriched epimastigote extract 25 ug + Ac 1 ug + probe UUUCUU 10 ng, lane 3 – Free probe (AAACAA) 4 – Enriched epimastigote extract 25 ug + Ac 1 ug + probe AAACAA 10 ng.
Discussion
There are two major classes of RNA granules in eukaryotes: stress granules and P-bodies. Stress granules (SGs) are formed when eIF2α is phosphorylated by stress-activated kinases, preventing new translation initiation events.Citation38 These granules appear under stress conditions and the core particle consists of translation initiation factors, such as eIF4E, eIF4A, eIF4G, eIF3, eIF2, the poly(A)-binding protein PABP1, and proteins from the small, but not the large ribosomal subunit. In mammalian cells, the SG components are nucleated by proteins such as TIA-1, TIAR and G3BP.Citation37-Citation40 In T. brucei, mammalian-like SGs appear in response to severe heat shock. They contain the poly(A)-binding proteins PABP1 and PABP2, all four isoforms of eIF4E (1 to 4), eIF2α, and eIF3B, but no ribosomal proteins.Citation41
Processing bodies (P-bodies) constitute the other class of RNA granules. They are constitutively present in cells and contain enzymes involved in mRNA decapping and degradation, such as the 5′ to 3′ exoribonuclease XRN1, the decapping complex Dcp1/Dcp2 and the Lsm1–7 protein complex in mammalian cells.Citation42 Ribosomal proteins are absent from P-bodies, although these granules contain translation initiation factors, such as eIF4E, eIF4G, and PABP1.Citation42 P-body-like structures have been described in both T. cruziCitation30,Citation31 and T. brucei.(43)
The protein composition of the TcZC3H39-mRNP included several ribosomal proteins, such as the 40S proteins S23, S29 and S30 in particular, and eukaryotic elongation and peptide release factor protein 1. In addition to translation-related proteins, we also identified ATP-dependent RNA helicase and RNA-binding proteins. The cellular distribution of TcZC3H39 changes over the life cycle of the parasite. In unstressed epimastigotes, this protein appears to be dispersed throughout the cytoplasm whereas, under nutritional stress conditions, it becomes slightly more granular. These different granule patterns are similar to what has been described for SGs and P-bodies, with a very conserved core and many proteins that interact dynamically with these granules. Furthermore, TcZC3H39 partially colocalizes with TcDHH1, a P-body marker. These differences in the structural organization of the TcZC3H39 mRNP may reflect differences in its function.
One particularly interesting observation was the association of the TcZC3H40 protein with TcZC3H39. The orthologs of these proteins in C. fasciculata, the cell cycle binding proteins CSBPA and CSBPB form a complex and bind cell cycle-regulated transcripts, stabilizing them via an octamer element in the 5′ UTR of the mRNA targets.Citation24,Citation25 In T. cruzi, these proteins also interact with each other to form the mRNP, as observed in C. fasciculata, but the targets are not the same as those described for CSBPA and CSBPB, as expected based on the evolutionary distance, reinforcing a different role for the TcZC3H39 and TcZC3H40 proteins in T. cruzi.
The identification of the mRNA targets associated with the TcZC3H39 protein made it possible to observe the dynamics of the RNA cycling occurring when T. cruzi epimastigotes are subjected to nutritional stress. This stress constitutes an important trigger for T. cruzi metacyclogenesis, leading to the transformation of non-infective epimastigotes into infective metacyclic trypomastigotes.Citation5,Citation44,Citation45 The analysis of the biological role assigned to the proteins encoded by the mRNAs associated with TcZC3H39, showed these proteins to be related to specific metabolic pathways in epimastigotes. However, mRNAs encoding other proteins, also related to specific functions in the cell, were observed associated with this protein in parasites subjected to nutritional stress. Thus, although there is a switch in targets, each set of targets consists of transcripts encoding proteins with related functions. During stress, the bound mRNAs encode proteins associated with mitochondria, such as subunits of the cytochrome c oxidase complex (COX) in particular, and ribosomal proteins. The metabolic activities of the cell (energetic metabolism and translation) are known to be decreased as an adaptive response to stress.Citation46 The association of mRNAs encoding proteins related to similar pathways with TcZC3H39 provides support for the posttranscriptional operon theory, according to which, related mRNAs are grouped into specific mRNPs to facilitate the control of their expression in the cell.Citation10 In T. brucei, the mRNAs encoding the components of oxoglutarate dehydrogenase and cytochrome oxidase subunits are co-regulated.Citation32,Citation34 These transcripts are more stable in the procyclic forms in which they are expressed to similar levels, whereas they are downregulated during differentiation into the bloodstream forms.Citation32,Citation34 This observation is consistent with our results showing an upregulation of transcript levels in unstressed epimastigotes and a drastic decrease in transcript levels during differentiation. TcZC3H39 probably plays a role in this modulation of mRNA expression.
In eukaryotes, the levels of the cytochrome c oxidase components (COX) are tightly controlled transcriptionally and posttranscriptionally.Citation47,Citation48 The posttranscriptional regulation of the COXIV protein involves the recognition of elements in the 3′-UTR of the transcript by the Hsp90 protein, leading to the inhibition of COXIV transcript translation.Citation49 In T. brucei, the COX complex constitutes a posttranscriptional operon and the trypanosome mitochondrial activity is downregulated in the bloodstream form.Citation32,Citation34 The instability of the COX transcripts is not due to the RNA silencing pathway, but to regulatory regions in the 3′-UTR regions of three COX mRNAs controlling the regulation of translation and transcript stability.Citation34,Citation50 In Saccharomyces cerevisiae, mitochondrial function is regulated in response to a shift between fermentable and non fermentable carbon sources, oxygen and heme.Citation51 The RNA-binding protein Puf3 binds 220 transcripts, most encoding mitochondrial proteins, and controls translation in the vicinity of the mitochondria.Citation51 In T. cruzi, an association of TcZC3H39 with COX V to IX transcripts and other nuclear-encoded mitochondrial mRNAs was observed, as previously demonstrated in T. brucei and S. cerevisiae. This association, which occurs in conditions of nutritional stress, suggests a potential role for the TcZC3H39-mRNP in mRNA storage. Consistent with this hypothesis, the mRNAs encoding COX components are less stable during stress conditions, but the decay rate is slow, indicating that these mRNAs are not being rapidly degraded. Similar results for COX transcript stability have also been obtained for T. brucei, in which the half-life in the procyclic form has been calculated to be four hours.Citation34
It is well-known that cells decrease their metabolism under stress conditions, and this is particularly true for energy metabolism, such as oxidative phosphorylation.Citation46 RNA binding proteins might play a role in the modulation on this process. The identification of TcZC3H39 mRNA targets and protein composition might shed light on the mechanisms involved in parasite stress responses and in the regulation of gene expression during T. cruzi differentiation.
Material and Methods
Parasite cultures
T. cruzi Dm 28c epimastigotes were cultured in liver infusion tryptose (LIT) medium at 28 °C. Epimastigotes in the late exponential growth phase were obtained from five-day cultures (density of 5 × 107 parasites.ml−1). For stress conditions, epimastigotes from five-day cultures were harvested by centrifugation at 7,000 × g for 5 min at 10 °C and incubated for 2 h at 28 °C in TAU medium (190 mM NaCl, 17 mM KCl, 2 mM MgCl2, 2 mM CaCl2, 8 mM phosphate buffer pH 6.0) at a density of 5 × 108 parasites.ml−1.
Phylogenetic and molecular evolutionary analyses
Phylogenetic and molecular evolutionary analyses were conducted with MEGA version 5.Citation52 We used ClustalW, with the default configuration for sequence alignment, and the phylogenetic tree was constructed by the neighbor-joining method. The evolutionary distances are expressed as the number of amino-acid substitutions per site. Bootstrap percentages (for 1000 replicas) are shown above the branches (when > 50).
Immunofluorescence and imaging
Immunofluorescence assays were performed as previously described.Citation30,Citation31 The images and the deconvolution were performed in Leica AF6000 Modular Systems with LAS AF 3.x. For the granules count the ImageJ v. 1.47 program was used. n = 25, ***p-value < 0.05.
TcZC3H39 immunoprecipitation assays: protein content
Immunoprecipitation assays were performed with the anti-TcZC3H39 antibody and cytoplasmic extracts from late exponential growth phase epimastigotes and epimastigotes subjected to nutritional stress. Mouse anti-TcZC3H39 antibody (15 µl) was incubated with 50 µl of goat anti-mouse magnetic beads (New England Biolabs) for 2 h at room temperature, with moderate stirring. Pre-immune serum was incubated with beads under the same conditions and used as a control for the specificity of immunoprecipitation reactions. After incubation, the resin was collected, the supernatant was discarded and the resin was then washed twice with PBS. All proteomics and ribonomics experiments were performed in triplicate and a pool of pre-immune sera was used as a control.
We obtained T. cruzi cytoplasmic extracts, by washing 1 x 109 parasites with PBS and lysing them in 2 ml of isotonic buffer (100 mM KCl, 5 mM MgCl2, 10 mM Hepes pH 7.0, protease inhibitor (1:100) and 200 U/mL RNase OUT) and subjected to lysis at 2,000 psi for 30 min at 4 °C. Cytoplasmic extracts were obtained by centrifugation at 10,000 x g for 10 min at 4 °C and incubated with the beads coupled to anti-TcZC3H39 antibody, the TcOlla antibodyCitation32 or the pre-immune serum, as described above, for 2 h at room temperature, with moderate shaking. For the RNase treatment, after lysis the extract was incubated with RNase A (10 μg ml−1) for 1 h at 37 °C prior to immunoprecipitation.
The immunoprecipitated complexes (IP) were collected and the supernatants (SP) were saved. The beads were washed three times with buffer IMP2 (100 mM KCl, 5 mM MgCl2, 10 mM Hepes pH 7.0, protease inhibitor (1:100) and 200 U/ml RNase OUT). The proteins linked to the beads were eluted with 150 µl glycine (0.1 M pH 2.0) and the pH was adjusted to 7.5 – 8.0. We then added 150 µl of proteomic buffer (6 M urea/2 M thiourea in 10 mM HEPES (pH 8.0)) to the samples. The proteins were reduced with 1mM dithiothreitol for 30 min, alkylated with 5.5 mM iodoacetamide for 20 min protected from light. Then, four volumes of 20 mM ammonium hydrogen carbonate were added. The digestion was performed for 16 h with trypsin (Promega) at a protease/protein ratio of 1/50. Tryptic peptides were purified using home made RP-C18 StageTip columns prior to MS analysis.Citation53
Peptide mixtures were separated by online reversed-phase (RP) nanoscale capillary liquid chromatography (nanoLC) and analyzed by electrospray tandem mass spectrometry (ESI MS/MS). The experiments were performed with a nanoLC 1D plus System (Eksigent, Dublin, CA) connected to the LTQ Orbitrap XL ETD mass spectrometer (mass spectrometry facility RPT02H PDTIS / Carlos Chagas Institute - Fiocruz Parana) equipped with a nanoelectrospray ion source (Thermo Scientific). Chromatographic separation of the peptides took place in a 15 cm fused silica emitter (75 µm inner diameter) in-house packed with reversed-phase ReproSil-Pur C18-AQ 3 µm resin (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany).
Peptide mixtures were injected onto the column with a flow rate of 250 nL/min and subsequently eluted from 5 to 40% acetonitrile in 0.1% formic acid in a 120 min gradient. The mass spectrometer was operated in data-dependent mode to automatically switch between MS and MS/MS (MS2) acquisition. Survey full-scan MS spectra (at 350 – 1650 m/z range) were acquired in the Orbitrap analyzer with resolution R = 60,000 at m/z 400 (after accumulation to a target value of 1,000,000 in the linear ion trap). The ten most intense ions were sequentially isolated and fragmented in the linear ion trap using collision-induced dissociation at a target value of 10,000. Former target ions selected for MS/MS were dynamically excluded for 90 s. Total cycle time was approximately 3 s. The general mass spectrometric conditions were: spray voltage, 2.4 kV; no sheath and auxiliary gas flow; ion transfer tube temperature, 100 °C; collision gas pressure, 1.3 mTorr; normalized collision energy using wide-band activation mode 35% for MS2. Ion selection thresholds were 250 counts for MS2. An activation q = 0.25 and activation time of 30 ms was applied in MS2 acquisitions. The “lock mass” option was enabled in all full scans to improve mass accuracy of precursor ions.Citation54
For western blot analysis, IP samples were separated by SDS-PAGE in a 15% polyacrylamide gel and the bands obtained were transferred to a nitrocellulose membrane. Nonspecific binding sites were blocked by incubating the membrane with 5% non-fat milk powder and 0.1% Tween-20 in PBS for 30 min. The membrane was incubated for 1 h with specific antibodies against TcZC3H39, S7, P0 or Dhh1. It was then thoroughly washed in PBS and incubated with goat alkaline phosphatase-conjugated anti-rabbit IgG (Sigma) diluted 1:10,000. The color reaction was developed with 5-bromo-4-chloro-3-indozyl phosphate and nitroblue tetrazolium (Promega).
Data analysis
Peak list picking, protein identification, quantification, and validation were done using the MaxQuant platform (version 1.3.0.5).Citation55 Database searching with fragment mass spectra was performed using the algorithm AndromedaCitation56 that is integrated into the MaxQuant environment. Protein identification was based on T. cruzi protein sequence databases (Esmeraldo-like, Non Esmeraldo-like, and Unassigned contigs, version 4.2 from August 15, 2012 available at TriTrypDBCitation57). A “decoy” approach, to measure the identification error, was used by reversing the sequence of each entry of the regular sequences and complemented with frequently observed contaminants (porcine trypsin, Achromobacter lyticus lysyl endopeptidase, and human keratins) and their reversed sequences. Search parameters specified a MS tolerance of 7 ppm, a MS/MS tolerance of 0.5 Da, full trypsin specificity, allowing up to two missed cleavages, allowing 1% error for protein and peptide identification calculated independently. Carbamidomethylation of cysteine was set as a fixed modification and oxidation of methionines and N-terminal acetylation (protein) were allowed as variable modifications. For validation of the identifications, a minimum of six amino-acids for peptide length and two peptides per protein were required. In addition, a false discovery rate (FDR) threshold of 0.01 was applied at both peptide and protein levels.
The expression analysis was performed using R environment through Limma package that uses linear models for the assessment of differential expression.Citation58 After analysis, the p-values were adjusted using the Benjamini–Hochberg (BH) methodCitation59 to control FDR. Only the proteins with FDR equal or lower than 0.2 were selected for further analysis. To select the proteins overrepresented at the imunoprecipitation of TcZC3H39, three comparisons were performed: 1) Immunoprecipitation using pre immunization serum, 2) Proteomic analyses of total extract from epimastigote formsCitation35 and 3) Immunoprecipitation using TcOlla serum,Citation32 a non related protein. Only proteins found enriched in the TcZC3H39 immunoprecipitation were considered for the analysis.
TcZC3H39 immunoprecipitation assays: ribonomics
For the identification of the mRNAs associated with the TcZC3H39 protein/complexes, mouse anti-TcZC3H39 antibody (15 µl) was incubated with 50 µl of goat anti-mouse magnetic beads (New England Biolabs) and 40 U/ml RNase OUT (Invitrogen) for 16 h at 4 °C, with shaking. Cytoplasmic extract, corresponding to 1 x 109 cells, was incubated with the beads, which had previously been linked to anti-TcZC3H39 antibody by incubation for 2 h at 4 °C, with shaking. The beads were washed three times with IMP1 buffer. The immunoprecipitated RNAs were eluted and purified with the RNeasy® (Qiagen) kit, with the “Animal Cells I” protocol of the manufacturer’s manual, with an additional column-based DNase treatment step. The purified RNAs were subjected to deep sequencing with the SOLiD 3 platform.
In silico data analysis
The sequencing data obtained were analyzed using CLC Genomics Workbench© v 5.5.1. The reads were trimmed on the basis of quality, using a threshold phred score of 15. The T. cruzi reference genome used for mapping was obtained from the NCBI database (AAHK01) and the alignment was performed as follows: Additional upstream and downstream sequences of 100 bases; minimum number of reads, 10; maximum number of mismatches, 2; nonspecific match limit, -2; use of colorspace encoding. We selected possible targets of TcZC3H39 mRNP with the β binomial statistical test (Baggerly’s test),Citation60 with a P value-corrected FDR of ≤ 1% and a minimum of a 4-fold change with respect to the control (pre-immune serum) for significance.
RT-PCR
CDNA (cDNA) was synthesized from 1 µg of immunoprecipitated RNA, with 1 µl of 10 µM oligo-dT primer (USB Corporation, Cleveland, OH) and 1 µl of reverse transcriptase (IMPROM II, Promega, Fitchburg, WI), according to the manufacturers’ instructions. PCR was performed with 20 ng of cDNA as the template, 20 mM TRIS-HCl (pH 8.4), 10 pmol of primers, 2.5 mM MgCl2, 0.0625 mM dNTPs, and 1 unit Taq polymerase (Invitrogen). The oligonucleotide primer sets used for PCR are described in Table S5. The following program was used: initial denaturation at 95 °C for 15 min and 45 cycles of 95 °C for 15 s, 62 or 64 °C for 20 s and 72 °C for 45 s. All reactions were performed in triplicate. We subjected 10 µl of the RT-PCR products to electrophoresis in a 2% agarose gel, with ethidium bromide staining for visualization of the bands obtained. Images of the gel were obtained with UVP Bioimaging Systems.
Functional annotation and the search for regulatory elements
For Gene Ontology term enrichment analysis for the classification of transcripts and for Kegg pathway map analysis,Citation61 we used the DAVID Annotation tool (http://david.abcc.ncifcrf.gov).Citation62 For enrichment analysis and functional annotation, the statistical P value is based on the EASE score, a modified one-tailed Fisher’s exact test P value, for which a value of 0 represents perfect enrichment, and values of up to 0.05 are considered to indicate strong enrichment in the annotation categories considered. We searched for regulatory elements in the 3′ UTR of the transcripts with RSAT - Regulatory Search Analysis Tools version 1.168 (http://rsat.ulb.ac.be/).Citation63,Citation64 The following parameters were used for motif detection: detection of over-represented words – right-tailed test; oligomer length 6; background model and estimation method – Markov; Markov chain order – 2; pseudo-frequency – 0.01; pseudo-frequency per oligo – 2.44e-06.
Sucrose density gradient separation
T. cruzi polysomes were isolated on sucrose gradients. Cells (5 × 108) were incubated with 100 μg.ml-1 cycloheximide for 10 min. They were kept on ice for 5 min and then pelleted by centrifugation (7,000 x g for 5 min at 4 °C) and washed with cold TKM buffer (10 mM Tris, pH 7.4, 300 mM KCl and 10 mM MgCl2) supplemented with 100 μg.ml−1 cycloheximide. The cell pellet was resuspended in 900 μl TKM supplemented with 100 μg.ml−1 cycloheximide, 10 μg.ml−1 heparin, 10 μM E-64, and 1:100 EDTA-free protease cocktail (Roche). The suspension was transferred to a new tube containing 100 μl of lysis buffer (TKM supplemented with 10% (v/v) NP-40 and 2 M sucrose), and homogenized by repeated passages through a pipette. Lysis was monitored by phase-contrast microscopy. The lysate was centrifuged at 18,000 × g, at 4 °C, for 5 min. The cleared supernatant (500 μl; equivalent to 5 × 108 cells) was layered onto linear 15 to 55% sucrose density gradients prepared in TKM buffer supplemented with inhibitors (100 μg.ml-1 cycloheximide, 10 μM E-64, 1 mM PMSF, and 1 mg.ml−1 heparin) and centrifuged at 4 °C for 2 h at 365,000 × g in a Beckman SW41 rotor. After centrifugation, 500 μl fractions were collected with the ISCO gradient fractionation system.
Assessment of the amounts of the TcZC3H39 target mRNAs by quantitative PCR
Quantitative RT-PCR was performed according to MIQE criteria.Citation65 Assays were performed in triplicate with epimastigotes cultured for five days in normal conditions (EPI) and epimastigotes cultured for five days under nutritional stress (STR), treated with 10 µg/ml actinomycin D. Total RNA was harvested after 0, 15 and 30 min, and mRNA levels at each time point were normalized with respect to the corresponding level of the histone H2A (TcCLB.508321.21) transcript and the control used for comparison was the transcript that codes the hypothetical protein (TcCLB.511737.70). The probes used in the assay were: cytochrome c oxidase subunit VI (TcCLB.511145.10), cytochrome c oxidase subunit VIII (TcCLB.511389.110), the component protein of the cytochrome c oxidase complex (TcCLB.506221.70) and the L9 ribosomal protein (TcCLB.504181.10). Total cellular RNA was isolated with the RNeasy® Mini Kit (Qiagen), according to the manufacturer’s instructions. DNA was digested with 1 U DNase (Promega) per µg RNA. The reverse transcription of RNA (1 µg) was performed with the oligo(dT) primer (Invitrogen) and ImProm-II™ Reverse Transcription System (Promega), using the default protocol. Real-time PCR was performed in triplicate, with the SYBR green master mix (Applied Biosystems), on an AB7500 (Applied Biosystems, Invitrogen). Expression levels were calculated by Livak's method.Citation66 Table S1 lists the primers used for quantitative PCR.
EMSA
RNA electrophoresis mobility shift assay (RNA-EMSA) and supershift assays were performed. Cy5.5-labeled RNA 3′-UTR probes (AAACAA) and nonspecific (UUUCUU) probes were purchased from Midland Inc. For the supershift assay, we incubated 2 mg of unstressed or stressed epimastigote extract with oligo dT beads, to enrich the extracts in proteins associated with mRNA. After extensive washing and elution, 25 µg of the enriched extract was used for the supershift assay. The labeled probe (10 ng) was incubated with 25 µg of enriched extract and 1 µg of TcZC3H39 antibody at room temperature for 30 min in binding buffer (10 mM TRIS-HCl, pH 7.4, 10 mM KCl, 1 mM MgCl2, 1 mM dithiothreitol, 200 ng.ml−1 heparin, 100 mM spermidine and 50% glycerol). The terminated reactions were subjected to electrophoresis in a 5% native polyacrylamide gel in 0.5% Tris-borate-EDTA buffer at 10 V/cm2. The gel was scanned using Odyssey Infrared Imaging system CLx. The intensity used on 700 nm channel was equal to 8.
Additional material
Download Zip (3.9 MB)Acknowledgments
This work was supported by Conselho Nacional do Desenvolvimenmto Científico e Tecnológico (CNPq), Fundação Oswaldo Cruz (Fiocruz, PAPES), Fundação Araucária and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). The authors thank the Program for Technological Development in Tools for Health-PDTIS-FIOCRUZ for use of its facilities. SG is a research fellow from CNPq.
We thank Vanessa Huf Sane for excellent technical assistance, Dr. Bruno Dallagiovanna for helpful discussions, Dr. Stênio Perdigão Fragoso and Dr. Daniela Fiori Gradia for the TcOLLA data and Dr. Fabiola Barbieri Holetz for the TcDhh1 antibody.
References
- De Souza W. Basic cell biology of Trypanosoma cruzi.. Curr Pharm Des 2002; 8:269 - 85; http://dx.doi.org/10.2174/1381612023396276; PMID: 11860366
- Das A, Banday M, Bellofatto V. RNA polymerase transcription machinery in trypanosomes. Eukaryot Cell 2008; 7:429 - 34; http://dx.doi.org/10.1128/EC.00297-07; PMID: 17951525
- Ferreira LRP, Dossin FdeM, Ramos TC, Freymüller E, Schenkman S. Active transcription and ultrastructural changes during Trypanosoma cruzi metacyclogenesis. An Acad Bras Cienc 2008; 80:157 - 66; http://dx.doi.org/10.1590/S0001-37652008000100011; PMID: 18345384
- Palenchar JB, Bellofatto V. Gene transcription in trypanosomes. Mol Biochem Parasitol 2006; 146:135 - 41; http://dx.doi.org/10.1016/j.molbiopara.2005.12.008; PMID: 16427709
- Goldenberg S, Avila AR. Aspects of Trypanosoma cruzi stage differentiation. Adv Parasitol 2011; 75:285 - 305; http://dx.doi.org/10.1016/B978-0-12-385863-4.00013-7; PMID: 21820561
- Clayton C, Shapira M. Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol Biochem Parasitol 2007; 156:93 - 101; http://dx.doi.org/10.1016/j.molbiopara.2007.07.007; PMID: 17765983
- Avila AR, Dallagiovanna B, Yamada-Ogatta SF, Monteiro-Góes V, Fragoso SP, Krieger MA, Goldenberg S. Stage-specific gene expression during Trypanosoma cruzi metacyclogenesis. Genet Mol Res 2003; 2:159 - 68; PMID: 12917812
- Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett 2008; 582:1977 - 86; http://dx.doi.org/10.1016/j.febslet.2008.03.004; PMID: 18342629
- Paquin N, Chartrand P. Local regulation of mRNA translation: new insights from the bud. Trends Cell Biol 2008; 18:105 - 11; http://dx.doi.org/10.1016/j.tcb.2007.12.004; PMID: 18262421
- Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet 2007; 8:533 - 43; http://dx.doi.org/10.1038/nrg2111; PMID: 17572691
- Kramer S, Kimblin NC, Carrington M. Genome-wide in silico screen for CCCH-type zinc finger proteins of Trypanosoma brucei, Trypanosoma cruzi and Leishmania major.. BMC Genomics 2010; 11:283; http://dx.doi.org/10.1186/1471-2164-11-283; PMID: 20444260
- Mörking PA, Dallagiovanna BM, Foti L, Garat B, Picchi GF, Umaki AC, Probst CM, Krieger MA, Goldenberg S, Fragoso SP. TcZFP1: a CCCH zinc finger protein of Trypanosoma cruzi that binds poly-C oligoribonucleotides in vitro. Biochem Biophys Res Commun 2004; 319:169 - 77; http://dx.doi.org/10.1016/j.bbrc.2004.04.162; PMID: 15158457
- Mörking PA, Rampazzo RdeC, Walrad P, Probst CM, Soares MJ, Gradia DF, Pavoni DP, Krieger MA, Matthews K, Goldenberg S, et al. The zinc finger protein TcZFP2 binds target mRNAs enriched during Trypanosoma cruzi metacyclogenesis. Mem Inst Oswaldo Cruz 2012; 107:790 - 9; http://dx.doi.org/10.1590/S0074-02762012000600014; PMID: 22990970
- Caro F, Bercovich N, Atorrasagasti C, Levin MJ, Vázquez MP. Protein interactions within the TcZFP zinc finger family members of Trypanosoma cruzi: implications for their functions. Biochem Biophys Res Commun 2005; 333:1017 - 25; http://dx.doi.org/10.1016/j.bbrc.2005.06.007; PMID: 15964555
- Hendriks EF, Matthews KR. Disruption of the developmental programme of Trypanosoma brucei by genetic ablation of TbZFP1, a differentiation-enriched CCCH protein. Mol Microbiol 2005; 57:706 - 16; http://dx.doi.org/10.1111/j.1365-2958.2005.04679.x; PMID: 16045615
- Ouna BA, Stewart M, Helbig C, Clayton C. The Trypanosoma brucei CCCH zinc finger proteins ZC3H12 and ZC3H13. Mol Biochem Parasitol 2012; 183:184 - 8; http://dx.doi.org/10.1016/j.molbiopara.2012.02.006; PMID: 22366391
- Benz C, Mulindwa J, Ouna B, Clayton C. The Trypanosoma brucei zinc finger protein ZC3H18 is involved in differentiation. Mol Biochem Parasitol 2011; 177:148 - 51; http://dx.doi.org/10.1016/j.molbiopara.2011.02.007; PMID: 21354218
- Paterou A, Walrad P, Craddy P, Fenn K, Matthews K. Identification and stage-specific association with the translational apparatus of TbZFP3, a CCCH protein that promotes trypanosome life-cycle development. J Biol Chem 2006; 281:39002 - 13; http://dx.doi.org/10.1074/jbc.M604280200; PMID: 17043361
- Ling AS, Trotter JR, Hendriks EF. A zinc finger protein, TbZC3H20, stabilizes two developmentally regulated mRNAs in trypanosomes. J Biol Chem 2011; 286:20152 - 62; http://dx.doi.org/10.1074/jbc.M110.139261; PMID: 21467035
- Walrad PB, Capewell P, Fenn K, Matthews KR. The post-transcriptional trans-acting regulator, TbZFP3, co-ordinates transmission-stage enriched mRNAs in Trypanosoma brucei.. Nucleic Acids Res 2012; 40:2869 - 83; http://dx.doi.org/10.1093/nar/gkr1106; PMID: 22140102
- Goldenberg S, Salles JM, Contreras VT, Lima Franco MP, Katzin AM, Colli W, Morel CM. Characterization of messenger RNA from epimastigotes and metacyclic trypomastigotes of Trypanosoma cruzi.. FEBS Lett 1985; 180:265 - 70; http://dx.doi.org/10.1016/0014-5793(85)81083-8; PMID: 2857136
- Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol 2002; 3:195 - 205; http://dx.doi.org/10.1038/nrm760; PMID: 11994740
- Alves LR, Avila AR, Correa A, Holetz FB, Mansur FCB, Manque PA, de Menezes JP, Buck GA, Krieger MA, Goldenberg S. Proteomic analysis reveals the dynamic association of proteins with translated mRNAs in Trypanosoma cruzi.. Gene 2010; 452:72 - 8; http://dx.doi.org/10.1016/j.gene.2009.12.009; PMID: 20060445
- Mahmood R, Hines JC, Ray DS. Identification of cis and trans elements involved in the cell cycle regulation of multiple genes in Crithidia fasciculata.. Mol Cell Biol 1999; 19:6174 - 82; PMID: 10454564
- Mahmood R, Mittra B, Hines JC, Ray DS. Characterization of the Crithidia fasciculata mRNA cycling sequence binding proteins. Mol Cell Biol 2001; 21:4453 - 9; http://dx.doi.org/10.1128/MCB.21.14.4453-4459.2001; PMID: 11416125
- Christensen DE, Klevit RE. Dynamic interactions of proteins in complex networks: identifying the complete set of interacting E2s for functional investigation of E3-dependent protein ubiquitination. FEBS J 2009; 276:5381 - 9; http://dx.doi.org/10.1111/j.1742-4658.2009.07249.x; PMID: 19712108
- Anantharaman V, Koonin EV, Aravind L. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res 2002; 30:1427 - 64; http://dx.doi.org/10.1093/nar/30.7.1427; PMID: 11917006
- DeRenzo C, Reese KJ, Seydoux G. Exclusion of germ plasm proteins from somatic lineages by cullin-dependent degradation. Nature 2003; 424:685 - 9; http://dx.doi.org/10.1038/nature01887; PMID: 12894212
- Anderson P, Kedersha N. Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones 2002; 7:213 - 21; http://dx.doi.org/10.1379/1466-1268(2002)007<0213:VSTROE>2.0.CO;2; PMID: 12380690
- Holetz FB, Correa A, Avila AR, Nakamura CV, Krieger MA, Goldenberg S. Evidence of P-body-like structures in Trypanosoma cruzi.. Biochem Biophys Res Commun 2007; 356:1062 - 7; http://dx.doi.org/10.1016/j.bbrc.2007.03.104; PMID: 17399688
- Holetz FB, Alves LR, Probst CM, Dallagiovanna B, Marchini FK, Manque P, Buck G, Krieger MA, Correa A, Goldenberg S. Protein and mRNA content of TcDHH1-containing mRNPs in Trypanosoma cruzi.. FEBS J 2010; 277:3415 - 26; http://dx.doi.org/10.1111/j.1742-4658.2010.07747.x; PMID: 20629747
- Gradia DF, Rau K, Umaki AC, de Souza FS, Probst CM, Correa A, Holetz FB, Avila AR, Krieger MA, Goldenberg S, et al. Characterization of a novel Obg-like ATPase in the protozoan Trypanosoma cruzi. Int J Parasitol 2009; 39:49 - 58; http://dx.doi.org/10.1016/j.ijpara.2008.05.019; PMID: 18713637
- Queiroz R, Benz C, Fellenberg K, Hoheisel JD, Clayton C. Transcriptome analysis of differentiating trypanosomes reveals the existence of multiple post-transcriptional regulons. BMC Genomics 2009; 10:495; http://dx.doi.org/10.1186/1471-2164-10-495; PMID: 19857263
- Ouellette M, Papadopoulou B. Coordinated gene expression by post-transcriptional regulons in African trypanosomes. J Biol 2009; 8:100; http://dx.doi.org/10.1186/jbiol203; PMID: 20017896
- de Godoy LM, Marchini FK, Pavoni DP, Rampazzo RdeC, Probst CM, Goldenberg S, Krieger MA. Quantitative proteomics of Trypanosoma cruzi during metacyclogenesis. Proteomics 2012; 12:2694 - 703; http://dx.doi.org/10.1002/pmic.201200078; PMID: 22761176
- Mayho M, Fenn K, Craddy P, Crosthwaite S, Matthews K. Post-transcriptional control of nuclear-encoded cytochrome oxidase subunits in Trypanosoma brucei: evidence for genome-wide conservation of life-cycle stage-specific regulatory elements. Nucleic Acids Res 2006; 34:5312 - 24; http://dx.doi.org/10.1093/nar/gkl598; PMID: 17012283
- van Helden J, Rios AF, Collado-Vides J. Discovering regulatory elements in non-coding sequences by analysis of spaced dyads. Nucleic Acids Res 2000; 28:1808 - 18; http://dx.doi.org/10.1093/nar/28.8.1808; PMID: 10734201
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol 1999; 147:1431 - 42; http://dx.doi.org/10.1083/jcb.147.7.1431; PMID: 10613902
- Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell 2009; 36:932 - 41; http://dx.doi.org/10.1016/j.molcel.2009.11.020; PMID: 20064460
- Anderson P, Kedersha N. Stress granules. Curr Biol 2009; 19:R397 - 8; http://dx.doi.org/10.1016/j.cub.2009.03.013; PMID: 19467203
- Thomas MG, Loschi M, Desbats MA, Boccaccio GL. RNA granules: the good, the bad and the ugly. Cell Signal 2011; 23:324 - 34; http://dx.doi.org/10.1016/j.cellsig.2010.08.011; PMID: 20813183
- Cassola A, Frasch AC. An RNA recognition motif mediates the nucleocytoplasmic transport of a trypanosome RNA-binding protein. J Biol Chem 2009; 284:35015 - 28; http://dx.doi.org/10.1074/jbc.M109.031633; PMID: 19801539
- Hoyle NP, Castelli LM, Campbell SG, Holmes LE, Ashe MP. Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J Cell Biol 2007; 179:65 - 74; http://dx.doi.org/10.1083/jcb.200707010; PMID: 17908917
- Cassola A, De Gaudenzi JG, Frasch AC. Recruitment of mRNAs to cytoplasmic ribonucleoprotein granules in trypanosomes. Mol Microbiol 2007; 65:655 - 70; http://dx.doi.org/10.1111/j.1365-2958.2007.05833.x; PMID: 17635187
- Contreras VT, Salles JM, Thomas N, Morel CM, Goldenberg S. In vitro differentiation of Trypanosoma cruzi under chemically defined conditions. Mol Biochem Parasitol 1985; 16:315 - 27; http://dx.doi.org/10.1016/0166-6851(85)90073-8; PMID: 3903496
- Bonaldo MC, Souto-Padron T, de Souza W, Goldenberg S. Cell-substrate adhesion during Trypanosoma cruzi differentiation. J Cell Biol 1988; 106:1349 - 58; http://dx.doi.org/10.1083/jcb.106.4.1349; PMID: 3283152
- Manoli I, Alesci S, Blackman MR, Su YA, Rennert OM, Chrousos GP. Mitochondria as key components of the stress response. Trends Endocrinol Metab 2007; 18:190 - 8; http://dx.doi.org/10.1016/j.tem.2007.04.004; PMID: 17500006
- Lenka N, Vijayasarathy C, Mullick J, Avadhani NG. Structural organization and transcription regulation of nuclear genes encoding the mammalian cytochrome c oxidase complex. Prog Nucleic Acid Res Mol Biol 1998; 61:309 - 44; http://dx.doi.org/10.1016/S0079-6603(08)60830-2; PMID: 9752724
- Aschrafi A, Schwechter AD, Mameza MG, Natera-Naranjo O, Gioio AE, Kaplan BB. MicroRNA-338 regulates local cytochrome c oxidase IV mRNA levels and oxidative phosphorylation in the axons of sympathetic neurons. J Neurosci 2008; 28:12581 - 90; http://dx.doi.org/10.1523/JNEUROSCI.3338-08.2008; PMID: 19020050
- Cannino G, Ferruggia E, Rinaldi AM. Proteins participating to the post-transcriptional regulation of the mitochondrial cytochrome c oxidase subunit IV via elements located in the 3’UTR. Mitochondrion 2009; 9:471 - 80; http://dx.doi.org/10.1016/j.mito.2009.08.007; PMID: 19703590
- Gerber AP, Herschlag D, Brown PO. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol 2004; 2:E79; http://dx.doi.org/10.1371/journal.pbio.0020079; PMID: 15024427
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011; 28:2731 - 9; http://dx.doi.org/10.1093/molbev/msr121; PMID: 21546353
- Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem 2003; 75:663 - 70; http://dx.doi.org/10.1021/ac026117i; PMID: 12585499
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 2006; 127:635 - 48; http://dx.doi.org/10.1016/j.cell.2006.09.026; PMID: 17081983
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 2008; 26:1367 - 72; http://dx.doi.org/10.1038/nbt.1511; PMID: 19029910
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 2011; 10:1794 - 805; http://dx.doi.org/10.1021/pr101065j; PMID: 21254760
- Aslett M, Aurrecoechea C, Berriman M, Brestelli J, Brunk BP, Carrington M, Depledge DP, Fischer S, Gajria B, Gao X, et al. TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res 2010; 38:D457 - 62; http://dx.doi.org/10.1093/nar/gkp851; PMID: 19843604
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004; 3:e3; http://dx.doi.org/10.2202/1544-6115.1027; PMID: 16646809
- Benjamini, Y, Hochberg, Y. Controlling the False Discovery Rate: A practical and powerful approach to multiple testing. J R Stat Soc 1995; Series B,57:289–300.
- Baggerly KA, Deng L, Morris JS, Aldaz CM. Differential expression in SAGE: accounting for normal between-library variation. Bioinformatics 2003; 19:1477 - 83; http://dx.doi.org/10.1093/bioinformatics/btg173; PMID: 12912827
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000; 28:27 - 30; http://dx.doi.org/10.1093/nar/28.1.27; PMID: 10592173
- Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4:44 - 57; http://dx.doi.org/10.1038/nprot.2008.211; PMID: 19131956
- van Helden J, André B, Collado-Vides J. Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J Mol Biol 1998; 281:827 - 42; http://dx.doi.org/10.1006/jmbi.1998.1947; PMID: 9719638
- Thomas-Chollier M, Defrance M, Medina-Rivera A, Sand O, Herrmann C, Thieffry D, van Helden J. RSAT 2011: regulatory sequence analysis tools. Nucleic Acids Res 2011; 39:W86-91; http://dx.doi.org/10.1093/nar/gkr377; PMID: 21715389
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 2009; 55:611 - 22; http://dx.doi.org/10.1373/clinchem.2008.112797; PMID: 19246619
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) Method. Methods 2001; 25:402 - 8; http://dx.doi.org/10.1006/meth.2001.1262; PMID: 11846609
