Abstract
Asymmetric, motor-protein dependent transport of mRNAs and subsequent localized translation is an important mechanism of gene regulation. Due to the high complexity of such motile particles, our mechanistic understanding of mRNA localization is limited. Over the last two decades, ASH1 mRNA localization in budding yeast has served as comparably simple and accessible model system. Recent advances have helped to draw an increasingly clear picture on the molecular mechanisms governing ASH1 mRNA localization from its co-transcriptional birth to its delivery at the site of destination. These new insights help to better understand the requirement of initial nuclear mRNPs, the molecular basis of specific mRNA-cargo recognition via cis-acting RNA elements, the different stages of RNP biogenesis and reorganization, as well as activation of the motile activity upon cargo binding. We discuss these aspects in context of published findings from other model organisms.
Introduction
mRNA localization is a universal feature throughout eukaryotes with important roles in cell polarity and differentiation, as well as embryonic development and body axis determination (reviewed in refs. 1–3). In somatic tissues mRNA localization is required for the formation and function of highly differentiated cells like neurons, neuroglia and epidermal cells. In higher eukaryotes, several dozens of factors usually co-immunoprecipitate in localizing messenger ribonucleoprotein particles (mRNPs).Citation4-6 This complexity is much reduced in protozoans like fungi.Citation7 However, for most protozoan model organisms, including the filamentous plant pathogen Ustilago maydis, the number of core factors remains unknown (reviewed in refs. 3 and 8). Several localizing mRNPs in higher and lower eukaryotes associate with membranes and vesicles (reviewed in refs. 9 and 10). Also the reported numbers of specifically recognized RNA species vary dramatically from thousandsCitation11-13 to only several dozens.Citation14 Most likely these numbers extensively depend on the threshold for the definition of specific binding and usually drastically decrease when cross-validation is performed.Citation14 Obviously, a model system with a much lower complexity is needed to understand the mechanisms underlying mRNA localization.
In the past 20 y, budding yeast Saccharomyces cerevisiae has emerged as a particularly well-suited model system to understand basic principles of the assembly and function of mRNA-transport complexes (reviewed in refs. 9 and 15). In haploid yeast cells, asymmetric cell division results in progenies with opposing mating types of either MATa or MATα (reviewed in ref. 16). The mating type is altered in the mother cell by transcription of the site-specific endonuclease HO (Homothallic switching)Citation17 and an HO-dependent genomic recombination in the MAT gene locus. Since HO is only expressed in the mother cell, the mating type of the daughter cell remains unchanged.
In 1996 two concurrent studies showed that the protein Ash1p (asymmetric synthesis of HO) localizes preferentially to the daughter cell, where it inhibits HO transcription and thus mating-type switching.Citation18,19 A third concurrent study reported the identification of five so-called SHE genes (SWI5-dependent HO expression), whose protein products are required for the asymmetric distribution of Ash1p.Citation20 Only one year later two laboratories published the surprising finding that the ASH1 transcript and not its protein product is actively transported into the daughter cell.Citation21,22 The locally restricted translation of the ASH1 mRNA causes the Ash1p-dependent repression of HO transcription in the daughter cell, thereby leading to the selective inhibition of mating-type switching (reviewed in ref. 16).
The SHE1 gene identified in a genetic screen by Jansen and colleagues encodes the type V myosin (MyoV) motor Myo4p.Citation20 MyoV motors constitute the main group of myosins for cargo transport (reviewed in ref. 23). The N-terminus consists of an ATP-hydrolyzing motor domain, followed by a calmodulin-bound lever arm, a coiled-coil dimerization domain, and a globular cargo-binding domain at its C-terminus. Dimeric MyoV binds to and moves processively along actin filaments (reviewed in ref. 23). She2p is an ASH1 mRNA-binding proteinCitation24-26 with a domain fold that lacks similarity to previously described RNA-binding domains.Citation27 She3p was initially shown to be an adaptor protein in the transport complex.Citation24,25,28 She4p/Dim1p belongs to the UNC-45/CRO/SHE4 (UCS)-domain containing protein family and binds to Myo4p and other myosins in yeast.Citation29,30 It has been suggested to be required for the structural integrity of Myo4pCitation29,30 and as determinant of Myo4p step-size on actin filaments.Citation31 However, its exact role is still not fully understood. She5p/Bni1p is a member of the formin family that is involved in the nucleation of linear actin filaments and thus required for cytoskeletal integrity during cell division (reviewed in ref. 32). Initial experiments already suggested that She2p is the core ASH1 mRNA-binding protein, which is bound by Myo4p-associated She3p to form an active transport complex ().Citation24,25,28
Figure 1. Schematic drawings of ASH1 mRNA localization in budding yeast. (A) Overview cartoon showing the major components of the transport complex and their sites of action. Left side indicates the mother cell, right side the daughter cell. (B) Directionality of ASH1 mRNP assembly is warranted by a gradient of affinity and specificity from the initial assembly of the co-transcriptional mRNP to the mature cytoplasmic transport complex. (C) Schematic representation of complex reorganization at the nuclear pore and at the bud tip. (D) Cartoon showing the stoichiometries and number of molecules assembling into a motile transport complex.Citation120 (E) Scheme of the ER association of the ASH1 mRNP and its sub-complexes. For a more detailed assessment of ER inheritance, please see the concomitant review by Singer-Krüger and Jansen.
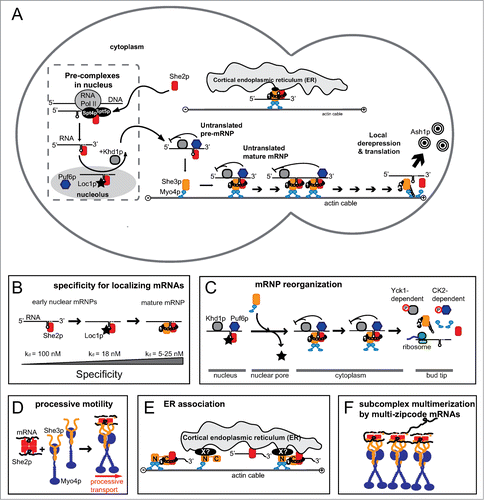
Besides these essential SHE factors, auxiliary proteins were discovered that also associate with the ASH1 mRNA. Their deletion resulted in the reduction, but not abolishment of ASH1 mRNA localization. These factors include the nucleolar ribosome biogenesis factor Loc1p, as well as the translational repressors Puf6p and Khd1p.Citation33-39 Aside from ASH1 mRNA, about 30 other transcripts are transported into the daughter cell.Citation7,Citation40-43
The assembly of the ASH1 mRNA-transport complex already occurs co-transcriptionally with the binding of the RNA-binding protein She2p to so-called localization or zip-code elements of the nascent ASH1 mRNA ( and ).Citation44 This core complex is joined by Loc1p, Puf6p, and Khd1p.Citation34-38,Citation45,46 While Loc1p remains in the nucleus, the rest of the ASH1 mRNP exports into the cytoplasm. There, She2p interacts with She3p ().Citation24,25,47 Since She3p itself is constitutively bound to the Myo4p motor,Citation24,25,Citation48-51 this interaction results in the formation of the mature transport complex. Together with its translational repressors, the ASH1 mRNP moves along actin filaments until it reaches the tip of the bud cell. At the bud tip, translation is activated. The result is Ash1p-dependent inhibition of the HO endonuclease and the prevention of mating-type switching in the daughter cell (reviewed in ref. 16).
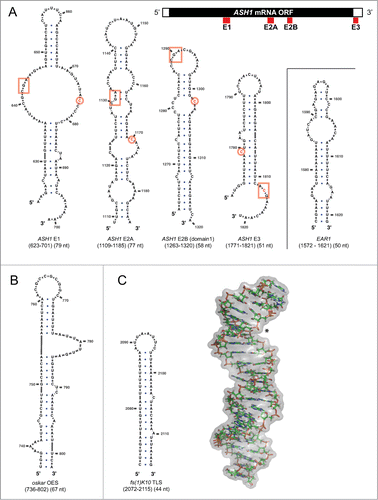
Figure 2. Secondary structures of zip-code elements (A) Selection of known zip-code elements bound by the She2p-She3p complex in yeast, as predicted by the mfold web server.Citation141 Cartoon on top shows that in the ASH1 mRNA, the zip-code elements E1, E2A, and E2B are part of the open reading frame, whereas E3 is located directly 3′ to the stop codon. Also the EAR1 mRNA contains an experimentally confirmed zip-code element. In the secondary structure predictions, boxed base triplets and a single encircled cytosine mark a previously identified consensus recognition motif.Citation68 Note that the EAR1 zip-code lacks this motif, that the motif can occur in inversed 3′ to 5′orientation, and that the base triplet is also found in permutations of its sequence. Also the location of the bipartite motif with regard to the secondary structure varies, indicating that the basic features of specific zip-code recognition are still not well understood. (B) OES secondary structure from chemical probing data.Citation62,76 (C) Secondary structure (left) and three-dimensional NMR structure (right) of fs(1)K10 TLS (PDB-identifier: 2KE6).Citation77 Asterisk shows widened major grove that is required for dynein-dependent localization.
Although the initial genetic screen did not offer any link to membrane transport, a subsequent screen revealed that two components of the mRNA transport complex, Myo4p and She3p are also required for the early inheritance of cortical endoplasmic reticulum (ER) to the bud cell ().Citation52 Vice versa, a subset of proteins required for cortical ER inheritance were also shown to affect mRNA localization,Citation43,53 suggesting a direct link between both processes.
While a concomitant review by Singer-Krüger and Jansen in this issue gives an overview on all the different mRNA transport processes present in yeast, this review focuses on recent advances of ASH1 mRNP function. In the remaining part of this review, we summarize the major progress made in the last five years to understand (1) why localizing mRNAs and transport-core factors are already assembled in mRNPs within the nucleus, (2) how specificity for localizing mRNAs is achieved, (3) what importance has mRNP reorganization at specific cellular sites has, (4) how processive motility of the ASH1 mRNP is achieved, (5) how ER inheritance is connected to mRNA localization, and (6) what effect multimerization of sub-complexes has on the function of localizing mRNPs.
Recent Advancements in Understanding ASH1 mRNP Function
Why do we need initial nuclear mRNPs?
Although the assembly of nuclear sub-complexes with localizing RNAs has been described in different organisms, to date it is not well understood why the nuclear history of mRNPs is important for cytoplasmic mRNA localization. Also in the budding yeast such initial nuclear complexes have been described. The core RNA-binding protein She2p shuttles between nucleus and cytoplasm.Citation48 An RNA-binding deficient version of She2p accumulates in the nucleolus,Citation45 indicating that She2p is exported with its target mRNA into the cytoplasm. Consistently, blocking nuclear mRNA export with a temperature-sensitive mutant of the export factor MEX67 traps She2p in the nucleus.Citation48 In addition, mutant versions of She2p, which are unable to enter the nucleus, show impaired cytoplasmic mRNA localization and upregulation of ASH1 mRNA translation.Citation45,46 Together these findings indicate that nuclear mRNA binding of She2p is required to mediate cytoplasmic inhibition of translation during transport.
More recently, it was shown that this nuclear association of She2p with ASH1 mRNA occurs already co-transcriptionally via binding to an RNA-polymerase II-associated heterodimeric protein complex, consisting of Suppressor of Ty's 4 and 5 (Spt4/5).Citation44 Based on these data Chartrand and colleagues proposed a model, in which She2p first associates with RNA polymerase II at the ASH1 locus and then selectively translocates onto the nascent ASH1 transcript to form a first, specific nuclear mRNP ().
For this intriguing model an open question concerns the specificity of co-transcriptional She2p recruitment to the ASH1 locus. In a second study a very similar approach was used to assess the association of She2p with chromatin and nascent RNA. However, it failed to confirm the previously reported selective enrichment of She2p at the ASH1 locus, but rather showed a broad, unspecific enrichment of She2p at chromatin.Citation47 Although it would make sense to selectively tether She2p co-transcriptionally to localizing mRNAs, it seems to require additional experimental proof to adequately understand these events.
A related unresolved issue is the association of She2p with the transcription machinery and its implications on specificity. The Spt4/5 complex acts as general RNA polymerase II-dependent elongation factor.Citation54,55 In contrast, She2p is selectively required for ASH1 mRNA recognition and localization. Considering that RNA-polymerase II is much more abundant in cells than She2pCitation56 and that RNA-polymerase II as well as the Spt4/5 complexes associate with a large number of loci, it remains unclear how She2p would efficiently find the few transcription sites of localizing mRNAs. Thus, more work seems to be required to fully understand how the She2p-dependent co-transcriptional recruitment results in the formation of specific ASH1 mRNPs.
In the nucleus this complex is joined by the translational repressors Puf6p and Khd1p ().Citation34-37 In experiments using a She2p mutant that lacks a nuclear localization signal less ASH1 mRNA is associated with Puf6p. Thus, She2p seems to be important for the formation of nuclear Puf6p-containing mRNPs.Citation46 Consistent with this finding, in cells with a cytoplasmically tethered mutant of She2p, translational repression is impaired during transport.Citation45 The third factor interacting with the ASH1 mRNA in the nucleus is Loc1p ().Citation38,45 This protein contains a number of potential nucleolar localization signals and directly interacts with She2p in a specific manner.Citation57 Furthermore, the incorporation of Loc1p into the ASH1 mRNA-She2p complex enhances the specificity and stability in vitro.Citation57 Loc1p is therefore likely to stabilize and direct the ASH1-She2p mRNP into the nucleolus, where it is loaded with the translational repressor Puf6p ().Citation36,37
Also examples from higher eukaryotes have been described, where initial nuclear mRNPs are important for cytoplasmic mRNA localization. For instance, neuronal Staufen2 is implicated in cytoplasmic RNA localization to dendrites, thereby likely modulating synaptic plasticity.Citation58 In similarity to studies with She2p, a mutant of Staufen2 with deficiency in RNA binding accumulates in the nucleolus.Citation59 This finding suggests the formation of initial nuclear mRNPs and co-export of Staufen2 with its target mRNA into the cytoplasm.
A mechanistically very interesting and well understood example is the requirement of nuclear splicing of oskar mRNA in the oocyte of the fruit fly Drosophila melanogaster for its cytoplasmic localization to the posterior pole.Citation60 For correct RNA localization two structural elements within the RNA are prerequisite: the spliced oskar localization element (SOLE)Citation61 and the oskar 3′UTR containing a dimerization element ().Citation62 In the unspliced oskar pre-mRNA two halves of a bipartite zip-code element are separated by an intron. The splicing reaction at the first exon-exon junction joins both halves, and thus creates this so-called SOLE. Upon splicing the exon junction complex (EJC) is deposited on the mRNA and contributes to the correct localization of the RNA to the posterior pole.Citation61
Another example for nuclear priming of mRNPs can be found in β-actin mRNA localization, which facilitates neurite outgrowth and cell migration. β-actin mRNA is already bound co-transcriptionally by its dedicated RNA-binding protein and localization factor zipcode-binding protein 1 (ZBP1). This nuclear recruitment of ZBP1 to β-actin mRNA depends on ZBP2.Citation63,64 Since a knock-down of ZBP2 inhibits neurite outgrowth, formation of these nuclear mRNPs seems to be a prerequisite for functional cytoplasmic β-actin mRNA localization.
Also the localization of Vgl1 and VegT mRNA in the African clawed frog Xenopus laevis requires nuclear mRNP assembly. Already in 2004, Kress and colleagues proposed the formation of a nuclear core complex consisting of heterogeneous nuclear ribonucleoprotein 1 (hnRNP), Vg1 RBP/vera, and zip-code containing mRNA that is necessary for mRNP localization to the vegetal pole of the oocyte.Citation65
Molecular Mechanisms of Specificity in mRNA Recognition
Cis-acting regions in localizing mRNAs: The zip-code elements
Localizing mRNAs contain cis-acting localization elements, also termed zip-code elements that are recognized by the transport machinery for their localization (reviewed in ref. 66). Zip-code elements are sufficient to mediate localization, even when placed into heterologous mRNAs.Citation67 In budding yeast, the ASH1 mRNA has four zip-code elements (). Three of them, termed E1, E2A, and E2B, reside in the coding region, whereas the most 3′-located E3 zip-code element is found directly after the stop-codon. Although all four elements are specifically bound by the SHE complex,Citation47 they show no obvious sequence conservation and great differences in their predicted secondary structures (). Khd1p binds to a region that includes the E1 element and Puf6p interacts with sequences in the E3 element.Citation34,36 This E3 zip-code element appears to play a particularly important role. While showing the highest affinity for She2p, it is the only element in the ASH1 mRNA that resides outside of the open reading frame (), and has a binding site for Puf6p.Citation26,36,67 Secondary structure predictions of all four zip-code elements indicate stem loops with mismatches like bulges and internal loops (). The only reported consensus between these elements is the presence of a CGA-base triplet and a single cytosine separated by six bases either 3′ or 5′ from the triplet.Citation68 While this small motif is found in all four elements of the ASH1 mRNA and also in the localizing RNAs IST2 and YMR171c, it is lacking in some other localizing RNAs, such as the EAR1 zip-code (). Furthermore, within the ASH1 zip-code elements, this motif is located in different secondary structures, can have an inverse 3′ to 5′ orientation, the sequence of its base triplet can be permutated, and the spacing of this bipartite motif seems variable (). For several of the other localizing mRNAs, no defined zip-code element has been identified to date. Thus, further work will be required to understand what structural and sequence features define a zip-code element for SHE-dependent localization.
Since the zip-code binding mode in budding yeast is not well understood, a comparison with zip-code elements in higher eukaryotes might offer common themes. For instance, it seems to be a general rule that zip-code elements consist of double-stranded regions with hairpins and well-defined mismatches (reviewed in ref. 69). Such mismatches appear to be important to promote specificity for protein interactions. An exception to this rule is the zip-code element β-actin mRNA,Citation70 which consists of a single-stranded bipartite recognition motif.Citation71,72
In particular in Drosophila a number of such zip-code elements has been identified for mRNAs expressed either during oogenesis or embryogenesis. They include the Gurken Localization Signal (GLS),Citation73 Bicoid Localization Element (BLE),Citation74,75 the above-described SOLE elementCitation61 and the Oocyte Entry Signal (OES)Citation76 of the oskar mRNA (), and a 44nt Transport/Localization Sequence (TLS) in the fs(1)K10 mRNA ().Citation77 Although these localizing mRNAs have distinct functions in development, at least some of them appear to have related zip-code elements. For instance, the fs(1)K10 mRNA normally localizes to the apical regions of the embryo. When its zip-code element TLS is expressed earlier during oogenesis as part of a heterologous RNA, this element also mediates its localization in its new environment.Citation77,78 A second example is the OES, which mediates the transport of oskar mRNA from the nurse cells into the oocyte.Citation76 When ectopically expressed, OES-containing RNAs are also localized in blastoderm-stage embryos, in follicular epithelial cells, and in larval salivary glands.Citation76 This observation indicates that the localization machinery is conserved and might be reused during different developmental stages of the fruit fly. Furthermore, fs(1)K10 TLS and the oskar OES are interchangeable.Citation76 This is surprising because both zip-code elements share no obvious homology on the level of sequence and secondary-structure predictions ( and ), thereby being very reminiscent of the ASH1 zip-codes ().
In comparison to double-stranded DNA binding motifs it is much more difficult to define consensus protein-binding motifs in RNAs. A main reason is that RNAs fold in a much more diverse manner, more similar to proteins than to DNA. As a consequence it is difficult to predict secondary or even tertiary structures with high reliability (reviewed in ref. 79). Thus, in most cases, secondary structure predictions and sequence similarities are insufficient to identify zip-code elements and to understand their function.
The example of the fs(1)K10 TLS from Drosophila shows that three-dimensional structural information are extremely useful to overcome this limitation. The 44nt long TLS zip-code element is a stem-loop structure with an A’-form helix.Citation77 This unusual helix has a widened major grove that is required for dynein-dependent localization to apical regions of the Drosophila embryo (). A second example where structural information are necessary to explain zip-code binding is ZBP1 from chicken and its RNA target β-actin mRNA. Here a 54nt long single-stranded zip-code element with a bipartite binding motif is necessary and sufficient for the localization of reporter mRNAs.Citation70-72 Important for specificity are not only the two binding motifs but also their exact spacing. Other interesting examples, where cis-acting targeting elements lead to transport of distinct mRNAs can be found in neurons. For instance the small non-coding brain cytoplasmic RNA 1 (BC1) is localized to dendrites of rodents,Citation80 as well as Mauthner axons and dendrites of goldfish,Citation81 where it regulates translation of target genes.Citation82,83 Dendritic localization elements can also be found in the α-calcium/calmodulin-dependent protein kinase II (α-CaMKII) mRNA. Here several cis-acting elements mediate site-directed transport.Citation84-88
RNA-Binding Domains for mRNA Recognition
It is interesting to note that only two of the RNA-binding proteins in the ASH1 mRNP bear known RNA-binding domains. The first one is the auxiliary factor Puf6p, which belongs to the highly conserved PUF (Pumilio and FBP) family of RNA-binding proteins. PUF proteins are characterized by a Pumilio-homology domain (PUM-HD).Citation89,90 The second is Khd1p, which contains three K-homology (KH) RNA-binding domains.Citation91,92 In contrast, no predictable RNA-binding domain can be found in the auxiliary factor Loc1p. Also the two zip-code recognizing RNA-binding proteins She2p and She3p lack any recognizable canonical RNA-binding domain.
A number of abundant RNA-binding domains have been identified in the past. Beside the above-described PUM-HD and the KH domains, other examples for wide-spread RNA-binding domains include the double-stranded RNA-binding domain (dsRBD),Citation93-95 the RNA-recognition motif (RRM),Citation96,97 zinc fingers (ZF),Citation98,99 PAZ domain,Citation100,101 PIWI domain,Citation102-104 and the Sterile alpha motif/pointed (SAM) domain.Citation105 Thus, at first glance this apparent lack of known domains in the ASH1 mRNP appears unusual.
However, recent studies suggest that also other RNA-binding proteins for mRNA localization lack identifiable RNA-binding domains. For instance, in Drosophila the RNA-binding protein Egalitarian shows no sequence similarity to known RNA-binding domains or motifs.Citation106 Furthermore, two recent systematic screens for mRNA-interacting proteins demonstrated that at least one-third of the identified factors lack identifiable RNA-binding domains and were previously not predicted to bind RNA.Citation107,108 These findings suggest that the RNA-interactome is larger than previously anticipated and that the high number of non-canonical RNA-binding proteins in the ASH1 mRNP might not be unusual after all.
Specific She2p-Loc1p mRNP in the Nucleus
In vitro binding experiments showed that the initial nuclear mRNP consisting of ASH1 mRNA and She2p has a low specificity and complex stability.Citation26,47 Both of these features seem incompatible with the idea that She2p stably and selectively escorts localizing mRNAs from their site of transcription to the final destination at the tip of the daughter cell. This discrepancy was resolved by the recent finding that the joining of Loc1p is able overcome this limitation. Loc1p binds simultaneously and directly to the ASH1 mRNA as well as to She2p, resulting in a ternary complex with much improved stability and specificity ().Citation57 Although it is unclear where exactly in the nucleus Loc1p joins the complex, it is reasonable to assume that this event should occur as early as possible, perhaps during or shortly after transcription. The requirement of the joint RNA-binding of two proteins is important also in subsequent stages of ASH1-mRNP function.
Such a cooperative RNP binding can also be found in the claw frog Xenopus laevis. Vg1 RBP/Vera, hnRNP1 and 40LoVe associate with Vg1 mRNA already in the nucleus and all factors affect vegetal mRNP localization.Citation109-111 40LoVe binds the localization element of the Vg1 mRNA (VLE) with high affinity but low specificity. Mutation or deletion of binding motifs of hnRNP1 and Vg1 RBP/Vera within the VLE abolishes also the binding of 40LoVe.Citation109,111 Therefore hnRNP1 and/or Vg1 RBP/Vera might be the recruitment factors that provide specificity for the localizing mRNAs.
Specific She2p-She3p mRNP in the Cytoplasm
Since no Loc1p is present in the cytoplasm, the exported She2p-ASH1 mRNA complex would be unstable in absence of another stabilizing co-factor. This problem is overcome by the joining of She3p. Besides being a myosin adaptor, the strictly cytoplasmic She3p is a rather unspecific RNA-binding protein that also undergoes protein-protein interactions with She2p.Citation47 The joining of She2p and She3p with ASH1 mRNA in the cytoplasm results in the formation of a highly specific, high-affinity co-complex with each of the four zip-code elements of the ASH1 mRNA ().Citation47 This synergistic complex together with its associated motor Myo4p and the translational repressors Puf6p and Khd1p constitutes the mature ASH1 transport complex. Interestingly, there is a gradual increase of binding affinity for the RNA from the co-transcriptional recruitment to the assembly of the mature cytoplasmic transport complex, ensuring directionality of assembly ().
From these recent insights on cargo-RNA binding in the nucleus and in the cytoplasm, it became clear that only the joint RNA binding of two proteins achieves specific zip-code recognition. In the nucleus She2p requires Loc1p and in the cytoplasm She3p takes over from Loc1p to form a highly specific ternary complex with She2p and zip-code RNA. These observations in yeast might serve as an emerging concept for zip-code recognition also in other organisms.
In the Drosophila embryo, the apical localization of fs(1)K10 and hairy mRNA depend on dynein-containing particles. This mRNP contains the two interacting proteins BicaudalD and Egalitarian, which in co-complex show higher affinity and specificity than the Egalitarian mRNP alone.Citation106 Although similar to the joint binding of She2p and She3p, this effect is less pronounced.
Recent studies on ZBP1 demonstrated how two of its KH domains mediate specificity for β-actin mRNA localization.Citation71,72 Chao and colleagues combined NMR data with a crystal structure to show that this zip-code element is single stranded with a bipartite recognition motif, which is recognized by two KH domains forming a defined di-domain.Citation71,72 The zip-code element wraps around this di-domain and binds at two opposing sites of the protein.Citation72 Whereas each binding motif interacts with one KH domain only with medium specificity, the spacer of defined length ensures the precise coordination of both binding events thus mediating specificity in RNA recognition.
Interestingly, the overwhelming majority of mRNA-binding proteins in higher eukaryotes have more than one RNA-binding domain (reviewed in ref. 112). For instance the protein Vigilin consists of 14 KH domains and the RNA-binding transcription factor III A (TFIIIA) contains nine zinc-finger motifs. Such proteins can also contain combinations of different types of RNA-binding domains, like the above-described ZBP1, which has four KH domains and two additional RRMs. It has been previously speculated that the combined action of two or more domains within one protein might be required to achieve high specificity.Citation112
Complex Reorganization at Nuclear Pore and at Bud Tip
The presence of a specific and stable nuclear mRNP indicates the need for reorganization processes to ensure the formation of the subsequent complex (e.g., mature cytoplasmic complex). Since Loc1p is a strictly nucle(ol)ar protein, it has to be removed from the ASH1 mRNP prior to its cytoplasmic transport. Indeed, in vitro pull-down experiments and UV-crosslinking studies demonstrated that the cytoplasmic co-complex of She2p and She3p out-completes and displaces Loc1p from the mRNP ( and ).Citation57 As Loc1p binds to both, the zip-code RNA and She2p, the out-competition by She3p must occur in a specific manner at the protein and the mRNA. She3p itself is strictly cytoplasmic and constitutively bound to the type V myosin Myo4p,Citation24,25,Citation48-51 whereas Loc1p has only been reported in the nucleus.Citation38,39,45 Thus Loc1p is likely removed from the complex close to the nuclear pore complex () during or directly after nuclear export. In cells with a genomic deletion of SHE3, the nuclear distribution of Loc1p is altered and a subfraction of Loc1p is observed in the cytoplasm, proving that complex remodeling at the nuclear pore depends on She3p.Citation57
Recently it was shown that phenylalanine-glycin repeat domains of yeast nucleoporins (Nup) are necessary for complex remodeling shortly after mRNP export. Nup159 and Nup42 position mRNPs for remodeling by the RNA helicase Dbp5, which triggers the release of mRNP from the transport receptor Mex67-Mtr2 and their export into the cytoplasm.Citation113 The dependence of nuclear export of She2p on Mex67 indicates that remodeling of the ASH1 mRNP at the nuclear pore might involve the same molecular pathway.
Once the cytoplasmic mRNPs are assembled, they move toward the plus ends of actin filaments at the bud tip, where translation is activated. About ten years ago it was shown that efficient ASH1 localization and localized translation involves complex reorganization at the bud tip.Citation114 This conclusion is consistent with the finding that proper localization at the bud tip requires the mRNA to bear a start codon,Citation34,115 suggesting that active translation might potentially be involved in anchoring.
More recently two studies demonstrated that both translational repressors, Khd1p and Puf6p, are phosphorylated at the bud tip, thereby reducing their affinity for the ASH1 mRNA ().Citation35,37 The posttranslational modifications result in reduced RNA binding, the release of ASH1 mRNA, and translation of the Ash1p. Phosphorylation-dependent release of localizing mRNAs at their site of destination has also been observed in higher eukaryotes.Citation116 Hence, it might serve as general principle for translational activation at the end of mRNA transport.
Understanding Motile Activity of Localizing ASH1 mRNPs
In vivo the ASH1 mRNP moves along actin filaments to the bud tip within 1–2 min,Citation117,118 where it is anchored.Citation119 Recently, in vitro reconstitution experiments with single-particle motility assays proved that the core factors Myo4p with its bound calmodulins and light chains, as well as She3p and She2p are sufficient to assemble a functional mRNA-transport complex.Citation120,121 Directional particle motility was also achieved in vitro when either cargo RNA was absent or when an RNA-binding deficient mutant of She2p was used for complex reconstitution.Citation120 Thus, the cargo RNA itself is dispensable for processive movement of the transport complex in vitro. Instead, the interaction of She2p with Myo4p-bound She3p activates the motile activity.Citation120
Unraveling the exact molecular composition of the ASH1 mRNP started already in 2007. At this time, it was reported that the Myo4p motor alone is monomeric and non-processive.Citation49,122,123 She3p binds to the rod of Myo4p and forms a single-headed myosin-She3p complex.Citation49,123 Other prerequisites for motile activity turned out to be the tetrameric state of She2pCitation26 and the need for at least two Myo4p molecules in one complex.Citation124 Subsequently, results from a cryo electron-microscopy study indeed showed that two Myo4p motors form a dimer-like structure in the particle.Citation125 Today the stoichiometric ratios and absolute numbers of molecules in a fully reconstituted and motile ASH1 mRNP are known.Citation120 One She2p tetramer binds two zip-code RNAs, thereby forming an initial nuclear complex. This pre-formed mRNP interacts with two complexes of a She3p dimer coupled to one Myo4p monomer.Citation120 Recently Shi and colleagues confirmed this She3p-Myo4p heterotrimer with the crystal structure of the C-terminal domain of Myo4p together with the N-terminal part of She3p.Citation51 The mature SHE core complex has a defined stoichiometric ratio, consisting of two ASH1 zip-codes, a She2p tetramer, two She3p dimers and two Myo4p monomers. This complex moves processively along actin filaments (Fig. 1A and D).Citation120 From these studies, one of the most surprising findings was the observation that actin transport complexes can also be assembled in absence of RNA.
In case of dynein-dependent transport in the Drosophila embryo, the RNA-cargo seems to play a more active role. Beside the protein levels of Egalitarian and Bicaudal D also the dosage of zip-code elements is responsible for a preferential minus-end-directed movement.Citation126-128
The ER Inheritance Connection
Interestingly, part of the localizing mRNA pool moves together with ER tubules to the bud.Citation53,129 Furthermore, some of the proteins required for cortical ER inheritance also affect the bud localization of a subset of mRNAs.Citation53 Because the majority of these co-migrating RNAs encode proteins involved in membrane biogenesis and function, their co-transport with ER implies a functional connection between ER and mRNA. Although She2p itself is dispensable for ER inheritance, its genomic deletion results in loss of localization of the above-described subset of mRNAs.Citation43,53 The recent observation that She2p directly associates with membranes independently from She3p or Myo4pCitation129,130 indicates that membrane-tethered She2p is responsible for binding of mRNAs to the cortical ER ().
In membrane-free ASH1 mRNPs, She3p is required for specific RNA recognition of She2p.Citation47 In the case of membrane-bound She2p it is still an unresolved issue whether She3p is also required for specific RNA-recognition. A number of recent studies suggest that such tethering of mRNAs to ER in a signal-recognition particle (SRP)-independent fashion is a rather common mechanism (reviewed in ref. 9 and 10). Since cortical ER is inherited into the daughter cell also in a SHE2-deficient strain,Citation52 a second open question remains how She3p and Myo4p are tethered to the ER (). Complex reconstitution experiments showed that She2p is required for Myo4p dimerization and is thus needed for processive movement (reviewed in ref. 15). It is therefore also unclear how Myo4p is activated in absence of She2p to mediate efficient ER inheritance ().
The Concept of Multimerization for mRNP Localization
In yeast, multiple zip-code elements exist in the ASH1 mRNA and in few other localizing transcripts.Citation24,41,42,115,117,131 Considering the observation that each motor-containing SHE complex contains two binding sites for zip-code elements,Citation120 every mRNA with multiple zip-codes should potentially be able to concatenate SHE complexes into larger particles. This hypothesis is supported by the in vivo observation that each transport particle contains several ASH1-mRNA molecules,Citation132,133 suggesting complex oligomerization also in the cell (). In vitro reconstitution experiments with RNAs bearing two zip-code elements confirmed that zip-code-mediated oligomerization of SHE complexes can occur.Citation120
In single particle motility assays Sladewski and colleagues found that mRNAs with multiple zip-code elements show increased processivity of movement.Citation121 While these particles were produced at non-stoichiometric ratios, a second study with mRNPs assembled with their correct stoichiometries did not find such zip code-dependent differences in motility.Citation120 Although the latter study suggests that there is no positive influence on processive movement caused by RNA-mediated oligomerization, the possibility exists that due to diverging subcellular localizations such strong sub-stoichiometric ratios could occur in the cell. However, due to the fact that stoichiometrically assembled ASH1 particles achieved run-lengths and velocitiesCitation120 comparable to in vivo observations,Citation67,118,119 it is unclear if a positive effect of multiple zip-codes on processivity would be required in vivo.
There could also be other reasons why multimerization by multi-zip code mRNAs is important for a yeast cell. For instance it could be advantageous to co-transport several mRNAs with a similar function. So-called post-transcriptional operons might be better co-regulated if they are transported in the same particles.Citation134,135 This interpretation would be beneficial for instance for the several mRNAs being associated with cortical ER. The majority of these mRNAs encode for proteins in membrane biogenesis and function.Citation53 Double-fluorescence staining in vivo indeed showed that two different RNAs can be co-transported in the same particle,Citation118 suggesting that co-regulation could indeed occur.
ASH1 mRNA is repressed during its transport by Puf6p and Khd1p.Citation34-37 For the other localizing mRNAs it is unclear, whether translational repression occurs by Puf6p and Khd1p. It is therefore tempting to speculate on translational repression of a subset of localizing mRNAs exclusively via the previously described multimerization of complexes into larger SHE particles.
The impact of mRNP mulitmerization has already been reported for oskar mRNA transport in Drosophila oocytes. The binding of the polypyrimidine tract-binding protein/hnRNP (PTB) to multiple sites of the oskar 3′UTR mediates the formation of higher-order RNPs and translational repression.Citation136 The 3′UTR of oskar mRNA also contains a dimerization element that interacts via kissing-loop between two RNA molecules and contributes to the enrichment of oskar mRNP at the posterior pole of the oocyte.Citation62 Such an RNA-mediated dimerization might also participate in the above described PTB-dependent translational repression of oskar mRNA.
RNA- and DNA-binding proteins show a significant enrichment of low-complexity (LC) regions when compared with the entire proteome.Citation137 For a long time, the functional implications of such low-complexity regions in RNA- and DNA-binding proteins were not well understood. A recent study found that (G/S-Y-G/S) motif-enriched LC regions in RNA-binding proteins like Fused in Sarcoma/Translocated in Sarcoma (FUS), Heterogeneous nuclear ribonucleoproteins A1 and 2 (hnRNPA1/2), Cytotoxic Granule-Associated RNA Binding Protein 1 (TIA1), Cytoplasmic polyadenylation element-binding protein 2 (CPEB2), or the Fragile X-Mental Retardation Protein (FMRP) can form reversible amyloid-like fibers in vitro.Citation138 In cells, the LC region of FUS was shown to be required for its localization to stress-granules.Citation138 Several of the RNA-binding proteins identified in this study are found in sub-cellularly localized mRNPs. Those include FMRP, IGF-II mRNA-binding proteins (IMP), HRP40, Barentz, Pumilio, and the hnRNP F/H homolog Glorund. Two subsequent studies demonstrated that the LC region of FUS mediates its RNA-dependent association with RNA polymerase II, indicating that RNA-dependent oligomerization might regulate transcriptional events.Citation139,140 It appears reasonable to assume that also in localizing mRNPs the LC regions of RNA-binding proteins play a functional role. They could offer a new organizing principle for oligomerization and assembly, and add an additional layer of regulation to this process.
Outlook
It is interesting to see that mechanistic key findings from yeast are conserved in other species. Synergistic RNA binding of zip-codes, the requirement of a nuclear history for proper cytoplasmic mRNA localization, complex reorganization at the nuclear pore and at the site of destination, as well as complex oligomerization of mRNPs are common principles found not only in yeast. Thus, this model organism will continue to provide essential information for understanding such general principles of mRNA localization.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We like to thank Birgit Singer-Krüger and Ralf-Peter Jansen for critical comments on this manuscript.
References
- Holt CE, Bullock SL. Subcellular mRNA localization in animal cells and why it matters. Science 2009; 326:1212-6; PMID:19965463; http://dx.doi.org/10.1126/science.1176488
- Jansen RP, Niessing D. Assembly of mRNA-protein complexes for directional mRNA transport in eukaryotes–an overview. Curr Protein Pept Sci 2012; 13:284-93; PMID:22708485; http://dx.doi.org/10.2174/138920312801619493
- Marchand V, Gaspar I, Ephrussi A. An intracellular transmission control protocol: assembly and transport of ribonucleoprotein complexes. Curr Opin Cell Biol 2012; 24:202-10; PMID:22278045; http://dx.doi.org/10.1016/j.ceb.2011.12.014
- Fritzsche R, Karra D, Bennett KL, Ang FY, Heraud-Farlow JE, Tolino M, Doyle M, Bauer KE, Thomas S, Planyavsky M, et al. Interactome of two diverse RNA granules links mRNA localization to translational repression in neurons. Cell Rep 2013; 5:1749-62; PMID:24360960; http://dx.doi.org/10.1016/j.celrep.2013.11.023
- Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron 2004; 43:513-25; PMID:15312650; http://dx.doi.org/10.1016/j.neuron.2004.07.022
- Maher-Laporte M, Berthiaume F, Moreau M, Julien LA, Lapointe G, Mourez M, DesGroseillers L. Molecular composition of staufen2-containing ribonucleoproteins in embryonic rat brain. PLoS One 2010; 5:e11350; PMID:20596529; http://dx.doi.org/10.1371/journal.pone.0011350
- Oeffinger M, Wei KE, Rogers R, DeGrasse JA, Chait BT, Aitchison JD, Rout MP. Comprehensive analysis of diverse ribonucleoprotein complexes. Nat Methods 2007; 4:951-6; PMID:17922018; http://dx.doi.org/10.1038/nmeth1101
- Göhre V, Haag C, Feldbrügge M. RNA biology in fungal phytopathogens. PLoS Pathog 2013; 9:e1003617; PMID:24146612; http://dx.doi.org/10.1371/journal.ppat.1003617
- Hermesh O, Jansen RP. Take the (RN)A-train: localization of mRNA to the endoplasmic reticulum. Biochim Biophys Acta 2013; 1833:2519-25; PMID:23353632; http://dx.doi.org/10.1016/j.bbamcr.2013.01.013
- Kraut-Cohen J, Gerst JE. Addressing mRNAs to the ER: cis sequences act up! Trends Biochem Sci 2010; 35:459-69; PMID:20346679; http://dx.doi.org/10.1016/j.tibs.2010.02.006
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr., Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 2010; 141:129-41; PMID:20371350; http://dx.doi.org/10.1016/j.cell.2010.03.009
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 2008; 456:464-9; PMID:18978773; http://dx.doi.org/10.1038/nature07488
- Maher-Laporte M, DesGroseillers L. Genome wide identification of Staufen2-bound mRNAs in embryonic rat brains. BMB Rep 2010; 43:344-8; PMID:20510018; http://dx.doi.org/10.5483/BMBRep.2010.43.5.344
- Heraud-Farlow JE, Sharangdhar T, Li X, Pfeifer P, Tauber S, Orozco D, Hörmann A, Thomas S, Bakosova A, Farlow AR, et al. Staufen2 regulates neuronal target RNAs. Cell Rep 2013; 5:1511-8; PMID:24360961; http://dx.doi.org/10.1016/j.celrep.2013.11.039
- Heym RG, Niessing D. Principles of mRNA transport in yeast. Cell Mol Life Sci 2012; 69:1843-53; PMID:22159587; http://dx.doi.org/10.1007/s00018-011-0902-4
- Cosma MP. Daughter-specific repression of Saccharomyces cerevisiae HO: Ash1 is the commander. EMBO Rep 2004; 5:953-7; PMID:15459746; http://dx.doi.org/10.1038/sj.embor.7400251
- Nasmyth K. Molecular analysis of a cell lineage. Nature 1983; 302:670-6; PMID:6339953; http://dx.doi.org/10.1038/302670a0
- Bobola N, Jansen RP, Shin TH, Nasmyth K. Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell 1996; 84:699-709; PMID:8625408; http://dx.doi.org/10.1016/S0092-8674(00)81048-X
- Sil A, Herskowitz I. Identification of asymmetrically localized determinant, Ash1p, required for lineage-specific transcription of the yeast HO gene. Cell 1996; 84:711-22; PMID:8625409; http://dx.doi.org/10.1016/S0092-8674(00)81049-1
- Jansen RP, Dowzer C, Michaelis C, Galova M, Nasmyth K. Mother cell-specific HO expression in budding yeast depends on the unconventional myosin myo4p and other cytoplasmic proteins. Cell 1996; 84:687-97; PMID:8625407; http://dx.doi.org/10.1016/S0092-8674(00)81047-8
- Long RM, Singer RH, Meng X, Gonzalez I, Nasmyth K, Jansen RP. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science 1997; 277:383-7; PMID:9219698; http://dx.doi.org/10.1126/science.277.5324.383
- Takizawa PA, Sil A, Swedlow JR, Herskowitz I, Vale RD. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature 1997; 389:90-3; PMID:9288973; http://dx.doi.org/10.1038/38015
- Trybus KM. Myosin V from head to tail. Cell Mol Life Sci 2008; 65:1378-89; PMID:18239852; http://dx.doi.org/10.1007/s00018-008-7507-6
- Böhl F, Kruse C, Frank A, Ferring D, Jansen RP. She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p. EMBO J 2000; 19:5514-24; PMID:11032818; http://dx.doi.org/10.1093/emboj/19.20.5514
- Long RM, Gu W, Lorimer E, Singer RH, Chartrand P. She2p is a novel RNA-binding protein that recruits the Myo4p-She3p complex to ASH1 mRNA. EMBO J 2000; 19:6592-601; PMID:11101531; http://dx.doi.org/10.1093/emboj/19.23.6592
- Müller M, Richter K, Heuck A, Kremmer E, Buchner J, Jansen RP, Niessing D. Formation of She2p tetramers is required for mRNA binding, mRNP assembly, and localization. RNA 2009; 15:2002-12; PMID:19710186; http://dx.doi.org/10.1261/rna.1753309
- Niessing D, Hüttelmaier S, Zenklusen D, Singer RH, Burley SK. She2p is a novel RNA binding protein with a basic helical hairpin motif. Cell 2004; 119:491-502; PMID:15537539; http://dx.doi.org/10.1016/j.cell.2004.10.018
- Takizawa PA, Vale RD. The myosin motor, Myo4p, binds Ash1 mRNA via the adapter protein, She3p. Proc Natl Acad Sci U S A 2000; 97:5273-8; PMID:10792032; http://dx.doi.org/10.1073/pnas.080585897
- Toi H, Fujimura-Kamada K, Irie K, Takai Y, Todo S, Tanaka K. She4p/Dim1p interacts with the motor domain of unconventional myosins in the budding yeast, Saccharomyces cerevisiae. Mol Biol Cell 2003; 14:2237-49; PMID:12808026; http://dx.doi.org/10.1091/mbc.E02-09-0616
- Wesche S, Arnold M, Jansen RP. The UCS domain protein She4p binds to myosin motor domains and is essential for class I and class V myosin function. Curr Biol 2003; 13:715-24; PMID:12725728; http://dx.doi.org/10.1016/S0960-9822(03)00264-1
- Shi H, Blobel G. UNC-45/CRO1/She4p (UCS) protein forms elongated dimer and joins two myosin heads near their actin binding region. Proc Natl Acad Sci U S A 2010; 107:21382-7; PMID:21115842; http://dx.doi.org/10.1073/pnas.1013038107
- Evangelista M, Zigmond S, Boone C. Formins: signaling effectors for assembly and polarization of actin filaments. J Cell Sci 2003; 116:2603-11; PMID:12775772; http://dx.doi.org/10.1242/jcs.00611
- Hasegawa Y, Irie K, Gerber AP. Distinct roles for Khd1p in the localization and expression of bud-localized mRNAs in yeast. RNA 2008; 14:2333-47; PMID:18805955; http://dx.doi.org/10.1261/rna.1016508
- Irie K, Tadauchi T, Takizawa PA, Vale RD, Matsumoto K, Herskowitz I. The Khd1 protein, which has three KH RNA-binding motifs, is required for proper localization of ASH1 mRNA in yeast. EMBO J 2002; 21:1158-67; PMID:11867544; http://dx.doi.org/10.1093/emboj/21.5.1158
- Paquin N, Ménade M, Poirier G, Donato D, Drouet E, Chartrand P. Local activation of yeast ASH1 mRNA translation through phosphorylation of Khd1p by the casein kinase Yck1p. Mol Cell 2007; 26:795-809; PMID:17588515; http://dx.doi.org/10.1016/j.molcel.2007.05.016
- Gu W, Deng Y, Zenklusen D, Singer RH. A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes Dev 2004; 18:1452-65; PMID:15198983; http://dx.doi.org/10.1101/gad.1189004
- Deng Y, Singer RH, Gu W. Translation of ASH1 mRNA is repressed by Puf6p-Fun12p/eIF5B interaction and released by CK2 phosphorylation. Genes Dev 2008; 22:1037-50; PMID:18413716; http://dx.doi.org/10.1101/gad.1611308
- Long RM, Gu W, Meng X, Gonsalvez G, Singer RH, Chartrand P. An exclusively nuclear RNA-binding protein affects asymmetric localization of ASH1 mRNA and Ash1p in yeast. J Cell Biol 2001; 153:307-18; PMID:11309412; http://dx.doi.org/10.1083/jcb.153.2.307
- Urbinati CR, Gonsalvez GB, Aris JP, Long RM. Loc1p is required for efficient assembly and nuclear export of the 60S ribosomal subunit. Mol Genet Genomics 2006; 276:369-77; PMID:16871394; http://dx.doi.org/10.1007/s00438-006-0151-7
- Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol 2008; 6:e255; PMID:18959479; http://dx.doi.org/10.1371/journal.pbio.0060255
- Shepard KA, Gerber AP, Jambhekar A, Takizawa PA, Brown PO, Herschlag D, DeRisi JL, Vale RD. Widespread cytoplasmic mRNA transport in yeast: identification of 22 bud-localized transcripts using DNA microarray analysis. Proc Natl Acad Sci U S A 2003; 100:11429-34; PMID:13679573; http://dx.doi.org/10.1073/pnas.2033246100
- Takizawa PA, DeRisi JL, Wilhelm JE, Vale RD. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science 2000; 290:341-4; PMID:11030653; http://dx.doi.org/10.1126/science.290.5490.341
- Aronov S, Gelin-Licht R, Zipor G, Haim L, Safran E, Gerst JE. mRNAs encoding polarity and exocytosis factors are cotransported with the cortical endoplasmic reticulum to the incipient bud in Saccharomyces cerevisiae. Mol Cell Biol 2007; 27:3441-55; PMID:17339339; http://dx.doi.org/10.1128/MCB.01643-06
- Shen Z, St-Denis A, Chartrand P. Cotranscriptional recruitment of She2p by RNA pol II elongation factor Spt4-Spt5/DSIF promotes mRNA localization to the yeast bud. Genes Dev 2010; 24:1914-26; PMID:20713510; http://dx.doi.org/10.1101/gad.1937510
- Du TG, Jellbauer S, Müller M, Schmid M, Niessing D, Jansen RP. Nuclear transit of the RNA-binding protein She2 is required for translational control of localized ASH1 mRNA. EMBO Rep 2008; 9:781-7; PMID:18566598; http://dx.doi.org/10.1038/embor.2008.112
- Shen Z, Paquin N, Forget A, Chartrand P. Nuclear shuttling of She2p couples ASH1 mRNA localization to its translational repression by recruiting Loc1p and Puf6p. Mol Biol Cell 2009; 20:2265-75; PMID:19244342; http://dx.doi.org/10.1091/mbc.E08-11-1151
- Müller M, Heym RG, Mayer A, Kramer K, Schmid M, Cramer P, Urlaub H, Jansen RP, Niessing D. A cytoplasmic complex mediates specific mRNA recognition and localization in yeast. PLoS Biol 2011; 9:e1000611; PMID:21526221; http://dx.doi.org/10.1371/journal.pbio.1000611
- Kruse C, Jaedicke A, Beaudouin J, Böhl F, Ferring D, Güttler T, Ellenberg J, Jansen RP. Ribonucleoprotein-dependent localization of the yeast class V myosin Myo4p. J Cell Biol 2002; 159:971-82; PMID:12499354; http://dx.doi.org/10.1083/jcb.200207101
- Heuck A, Du TG, Jellbauer S, Richter K, Kruse C, Jaklin S, Müller M, Buchner J, Jansen RP, Niessing D. Monomeric myosin V uses two binding regions for the assembly of stable translocation complexes. Proc Natl Acad Sci U S A 2007; 104:19778-83; PMID:18056806; http://dx.doi.org/10.1073/pnas.0706780104
- Heuck A, Fetka I, Brewer DN, Hüls D, Munson M, Jansen RP, Niessing D. The structure of the Myo4p globular tail and its function in ASH1 mRNA localization. J Cell Biol 2010; 189:497-510; PMID:20439999; http://dx.doi.org/10.1083/jcb.201002076
- Shi H, Singh N, Esselborn F, Blobel G. Structure of a myosin•adaptor complex and pairing by cargo. Proc Natl Acad Sci U S A 2014; 111:E1082-90; PMID:24522109; http://dx.doi.org/10.1073/pnas.1401428111
- Estrada P, Kim J, Coleman J, Walker L, Dunn B, Takizawa P, Novick P, Ferro-Novick S. Myo4p and She3p are required for cortical ER inheritance in Saccharomyces cerevisiae. J Cell Biol 2003; 163:1255-66; PMID:14691136; http://dx.doi.org/10.1083/jcb.200304030
- Fundakowski J, Hermesh O, Jansen RP. Localization of a subset of yeast mRNAs depends on inheritance of endoplasmic reticulum. Traffic 2012; 13:1642-52; PMID:22994588; http://dx.doi.org/10.1111/tra.12011
- Hartzog GA, Wada T, Handa H, Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev 1998; 12:357-69; PMID:9450930; http://dx.doi.org/10.1101/gad.12.3.357
- Mayer A, Lidschreiber M, Siebert M, Leike K, Söding J, Cramer P. Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol 2010; 17:1272-8; PMID:20818391; http://dx.doi.org/10.1038/nsmb.1903
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature 2003; 425:737-41; PMID:14562106; http://dx.doi.org/10.1038/nature02046
- Niedner A, Müller M, Moorthy BT, Jansen RP, Niessing D. Role of Loc1p in assembly and reorganization of nuclear ASH1 messenger ribonucleoprotein particles in yeast. Proc Natl Acad Sci U S A 2013; 110:E5049-58; PMID:24324176; http://dx.doi.org/10.1073/pnas.1315289111
- Doyle M, Kiebler MA. Mechanisms of dendritic mRNA transport and its role in synaptic tagging. EMBO J 2011; 30:3540-52; PMID:21878995; http://dx.doi.org/10.1038/emboj.2011.278
- Macchi P, Brownawell AM, Grunewald B, DesGroseillers L, Macara IG, Kiebler MA. The brain-specific double-stranded RNA-binding protein Staufen2: nucleolar accumulation and isoform-specific exportin-5-dependent export. J Biol Chem 2004; 279:31440-4; PMID:15166236; http://dx.doi.org/10.1074/jbc.C400226200
- Hachet O, Ephrussi A. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature 2004; 428:959-63; PMID:15118729; http://dx.doi.org/10.1038/nature02521
- Ghosh S, Marchand V, Gáspár I, Ephrussi A. Control of RNP motility and localization by a splicing-dependent structure in oskar mRNA. Nat Struct Mol Biol 2012; 19:441-9; PMID:22426546; http://dx.doi.org/10.1038/nsmb.2257
- Jambor H, Brunel C, Ephrussi A. Dimerization of oskar 3′ UTRs promotes hitchhiking for RNA localization in the Drosophila oocyte. RNA 2011; 17:2049-57; PMID:22028360; http://dx.doi.org/10.1261/rna.2686411
- Pan F, Hüttelmaier S, Singer RH, Gu W. ZBP2 facilitates binding of ZBP1 to beta-actin mRNA during transcription. Mol Cell Biol 2007; 27:8340-51; PMID:17893325; http://dx.doi.org/10.1128/MCB.00972-07
- Gu W, Pan F, Zhang H, Bassell GJ, Singer RH. A predominantly nuclear protein affecting cytoplasmic localization of beta-actin mRNA in fibroblasts and neurons. J Cell Biol 2002; 156:41-51; PMID:11781334; http://dx.doi.org/10.1083/jcb.200105133
- Kress TL, Yoon YJ, Mowry KL. Nuclear RNP complex assembly initiates cytoplasmic RNA localization. J Cell Biol 2004; 165:203-11; PMID:15096527; http://dx.doi.org/10.1083/jcb.200309145
- Singer RH. RNA zipcodes for cytoplasmic addresses. Curr Biol 1993; 3:719-21; PMID:15335871; http://dx.doi.org/10.1016/0960-9822(93)90079-4
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol Cell 1998; 2:437-45; PMID:9809065; http://dx.doi.org/10.1016/S1097-2765(00)80143-4
- Olivier C, Poirier G, Gendron P, Boisgontier A, Major F, Chartrand P. Identification of a conserved RNA motif essential for She2p recognition and mRNA localization to the yeast bud. Mol Cell Biol 2005; 25:4752-66; PMID:15899876; http://dx.doi.org/10.1128/MCB.25.11.4752-4766.2005
- Jambhekar A, Derisi JL. Cis-acting determinants of asymmetric, cytoplasmic RNA transport. RNA 2007; 13:625-42; PMID:17449729; http://dx.doi.org/10.1261/rna.262607
- Kislauskis EH, Zhu X, Singer RH. Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. J Cell Biol 1994; 127:441-51; PMID:7929587; http://dx.doi.org/10.1083/jcb.127.2.441
- Chao JA, Patskovsky Y, Patel V, Levy M, Almo SC, Singer RH. ZBP1 recognition of beta-actin zipcode induces RNA looping. Genes Dev 2010; 24:148-58; PMID:20080952; http://dx.doi.org/10.1101/gad.1862910
- Patel VL, Mitra S, Harris R, Buxbaum AR, Lionnet T, Brenowitz M, Girvin M, Levy M, Almo SC, Singer RH, et al. Spatial arrangement of an RNA zipcode identifies mRNAs under post-transcriptional control. Genes Dev 2012; 26:43-53; PMID:22215810; http://dx.doi.org/10.1101/gad.177428.111
- Van De Bor V, Hartswood E, Jones C, Finnegan D, Davis I. gurken and the I factor retrotransposon RNAs share common localization signals and machinery. Dev Cell 2005; 9:51-62; PMID:15992540; http://dx.doi.org/10.1016/j.devcel.2005.04.012
- Macdonald PM, Kerr K. Mutational analysis of an RNA recognition element that mediates localization of bicoid mRNA. Mol Cell Biol 1998; 18:3788-95; PMID:9632762
- Macdonald PM, Kerr K, Smith JL, Leask A. RNA regulatory element BLE1 directs the early steps of bicoid mRNA localization. Development 1993; 118:1233-43; PMID:8269850
- Jambor H, Mueller S, Bullock SL, Ephrussi A. A stem-loop structure directs oskar mRNA to microtubule minus ends. RNA 2014; 20:429-39; PMID:24572808; http://dx.doi.org/10.1261/rna.041566.113
- Bullock SL, Ringel I, Ish-Horowicz D, Lukavsky PJ. A’-form RNA helices are required for cytoplasmic mRNA transport in Drosophila. Nat Struct Mol Biol 2010; 17:703-9; PMID:20473315; http://dx.doi.org/10.1038/nsmb.1813
- Cohen RS, Zhang S, Dollar GL. The positional, structural, and sequence requirements of the Drosophila TLS RNA localization element. RNA 2005; 11:1017-29; PMID:15987813; http://dx.doi.org/10.1261/rna.7218905
- Weeks KM. Advances in RNA structure analysis by chemical probing. Curr Opin Struct Biol 2010; 20:295-304; PMID:20447823; http://dx.doi.org/10.1016/j.sbi.2010.04.001
- Muslimov IA, Santi E, Homel P, Perini S, Higgins D, Tiedge H. RNA transport in dendrites: a cis-acting targeting element is contained within neuronal BC1 RNA. J Neurosci 1997; 17:4722-33; PMID:9169532
- Muslimov IA, Titmus M, Koenig E, Tiedge H. Transport of Neuronal BC1 RNA in Mauthner Axons. J Neurosci 2002; 22:4293-301; PMID:12040034
- Eom T, Berardi V, Zhong J, Risuleo G, Tiedge H. Dual nature of translational control by regulatory BC RNAs. Mol Cell Biol 2011; 31:4538-49; PMID:21930783; http://dx.doi.org/10.1128/MCB.05885-11
- Lin D, Pestova TV, Hellen CU, Tiedge H. Translational control by a small RNA: dendritic BC1 RNA targets the eukaryotic initiation factor 4A helicase mechanism. Mol Cell Biol 2008; 28:3008-19; PMID:18316401; http://dx.doi.org/10.1128/MCB.01800-07
- Mayford M, Baranes D, Podsypanina K, Kandel ER. The 3′-untranslated region of CaMKII alpha is a cis-acting signal for the localization and translation of mRNA in dendrites. Proc Natl Acad Sci U S A 1996; 93:13250-5; PMID:8917577; http://dx.doi.org/10.1073/pnas.93.23.13250
- Mori Y, Imaizumi K, Katayama T, Yoneda T, Tohyama M. Two cis-acting elements in the 3′ untranslated region of alpha-CaMKII regulate its dendritic targeting. Nat Neurosci 2000; 3:1079-84; PMID:11036263; http://dx.doi.org/10.1038/80591
- Gao Y, Tatavarty V, Korza G, Levin MK, Carson JH. Multiplexed dendritic targeting of alpha calcium calmodulin-dependent protein kinase II, neurogranin, and activity-regulated cytoskeleton-associated protein RNAs by the A2 pathway. Mol Biol Cell 2008; 19:2311-27; PMID:18305102; http://dx.doi.org/10.1091/mbc.E07-09-0914
- Huang YS, Carson JH, Barbarese E, Richter JD. Facilitation of dendritic mRNA transport by CPEB. Genes Dev 2003; 17:638-53; PMID:12629046; http://dx.doi.org/10.1101/gad.1053003
- Tübing F, Vendra G, Mikl M, Macchi P, Thomas S, Kiebler MA. Dendritically localized transcripts are sorted into distinct ribonucleoprotein particles that display fast directional motility along dendrites of hippocampal neurons. J Neurosci 2010; 30:4160-70; PMID:20237286; http://dx.doi.org/10.1523/JNEUROSCI.3537-09.2010
- Wang X, McLachlan J, Zamore PD, Hall TM. Modular recognition of RNA by a human pumilio-homology domain. Cell 2002; 110:501-12; PMID:12202039; http://dx.doi.org/10.1016/S0092-8674(02)00873-5
- Wang X, Zamore PD, Hall TM. Crystal structure of a Pumilio homology domain. Mol Cell 2001; 7:855-65; PMID:11336708; http://dx.doi.org/10.1016/S1097-2765(01)00229-5
- Lewis HA, Musunuru K, Jensen KB, Edo C, Chen H, Darnell RB, Burley SK. Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell 2000; 100:323-32; PMID:10676814; http://dx.doi.org/10.1016/S0092-8674(00)80668-6
- Musco G, Stier G, Joseph C, Castiglione Morelli MA, Nilges M, Gibson TJ, Pastore A. Three-dimensional structure and stability of the KH domain: molecular insights into the fragile X syndrome. Cell 1996; 85:237-45; PMID:8612276; http://dx.doi.org/10.1016/S0092-8674(00)81100-9
- Bycroft M, Grünert S, Murzin AG, Proctor M, St Johnston D. NMR solution structure of a dsRNA binding domain from Drosophila staufen protein reveals homology to the N-terminal domain of ribosomal protein S5. EMBO J 1995; 14:3563-71; PMID:7628456
- Kharrat A, Macias MJ, Gibson TJ, Nilges M, Pastore A. Structure of the dsRNA binding domain of E. coli RNase III. EMBO J 1995; 14:3572-84; PMID:7628457
- Ryter JM, Schultz SC. Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J 1998; 17:7505-13; PMID:9857205; http://dx.doi.org/10.1093/emboj/17.24.7505
- Nagai K, Oubridge C, Jessen TH, Li J, Evans PR. Crystal structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein A. Nature 1990; 348:515-20; PMID:2147232; http://dx.doi.org/10.1038/348515a0
- Oubridge C, Ito N, Evans PR, Teo CH, Nagai K. Crystal structure at 1.92 A resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature 1994; 372:432-8; PMID:7984237; http://dx.doi.org/10.1038/372432a0
- De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science 1998; 279:384-8; PMID:9430589; http://dx.doi.org/10.1126/science.279.5349.384
- Lee MS, Gippert GP, Soman KV, Case DA, Wright PE. Three-dimensional solution structure of a single zinc finger DNA-binding domain. Science 1989; 245:635-7; PMID:2503871; http://dx.doi.org/10.1126/science.2503871
- Ma JB, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature 2004; 429:318-22; PMID:15152257; http://dx.doi.org/10.1038/nature02519
- Song JJ, Liu J, Tolia NH, Schneiderman J, Smith SK, Martienssen RA, Hannon GJ, Joshua-Tor L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat Struct Biol 2003; 10:1026-32; PMID:14625589; http://dx.doi.org/10.1038/nsb1016
- Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature 2005; 434:666-70; PMID:15800629; http://dx.doi.org/10.1038/nature03514
- Parker JS, Roe SM, Barford D. Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. EMBO J 2004; 23:4727-37; PMID:15565169; http://dx.doi.org/10.1038/sj.emboj.7600488
- Parker JS, Roe SM, Barford D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature 2005; 434:663-6; PMID:15800628; http://dx.doi.org/10.1038/nature03462
- Oberstrass FC, Lee A, Stefl R, Janis M, Chanfreau G, Allain FH. Shape-specific recognition in the structure of the Vts1p SAM domain with RNA. Nat Struct Mol Biol 2006; 13:160-7; PMID:16429156; http://dx.doi.org/10.1038/nsmb1038
- Dienstbier M, Boehl F, Li X, Bullock SL. Egalitarian is a selective RNA-binding protein linking mRNA localization signals to the dynein motor. Genes Dev 2009; 23:1546-58; PMID:19515976; http://dx.doi.org/10.1101/gad.531009
- Baltz AG, Munschauer M, Schwanhäusser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell 2012; 46:674-90; PMID:22681889; http://dx.doi.org/10.1016/j.molcel.2012.05.021
- Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 2012; 149:1393-406; PMID:22658674; http://dx.doi.org/10.1016/j.cell.2012.04.031
- Czaplinski K, Köcher T, Schelder M, Segref A, Wilm M, Mattaj IW. Identification of 40LoVe, a Xenopus hnRNP D family protein involved in localizing a TGF-beta-related mRNA during oogenesis. Dev Cell 2005; 8:505-15; PMID:15809033; http://dx.doi.org/10.1016/j.devcel.2005.01.012
- Czaplinski K, Mattaj IW. 40LoVe interacts with Vg1RBP/Vera and hnRNP I in binding the Vg1-localization element. RNA 2006; 12:213-22; PMID:16373488; http://dx.doi.org/10.1261/rna.2820106
- Kroll TT, Swenson LB, Hartland EI, Snedden DD, Goodson HV, Huber PW. Interactions of 40LoVe within the ribonucleoprotein complex that forms on the localization element of Xenopus Vg1 mRNA. Mech Dev 2009; 126:523-38; PMID:19345262; http://dx.doi.org/10.1016/j.mod.2009.03.007
- Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol 2007; 8:479-90; PMID:17473849; http://dx.doi.org/10.1038/nrm2178
- Adams RL, Terry LJ, Wente SR. Nucleoporin FG Domains Facilitate mRNP Remodeling at the Cytoplasmic Face of the Nuclear Pore Complex. [Epub ahead of print]. Genetics 2014; PMID:24931410; http://dx.doi.org/10.1534/genetics.114.164012
- Gonsalvez GB, Little JL, Long RM. ASH1 mRNA anchoring requires reorganization of the Myo4p-She3p-She2p transport complex. J Biol Chem 2004; 279:46286-94; PMID:15328357; http://dx.doi.org/10.1074/jbc.M406086200
- Gonzalez I, Buonomo SB, Nasmyth K, von Ahsen U. ASH1 mRNA localization in yeast involves multiple secondary structural elements and Ash1 protein translation. Curr Biol 1999; 9:337-40; PMID:10209099; http://dx.doi.org/10.1016/S0960-9822(99)80145-6
- Hüttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature 2005; 438:512-5; PMID:16306994; http://dx.doi.org/10.1038/nature04115
- Chartrand P, Meng XH, Hüttelmaier S, Donato D, Singer RH. Asymmetric sorting of ash1p in yeast results from inhibition of translation by localization elements in the mRNA. Mol Cell 2002; 10:1319-30; PMID:12504008; http://dx.doi.org/10.1016/S1097-2765(02)00694-9
- Lange S, Katayama Y, Schmid M, Burkacky O, Bräuchle C, Lamb DC, Jansen RP. Simultaneous transport of different localized mRNA species revealed by live-cell imaging. Traffic 2008; 9:1256-67; PMID:18485054; http://dx.doi.org/10.1111/j.1600-0854.2008.00763.x
- Beach DL, Salmon ED, Bloom K. Localization and anchoring of mRNA in budding yeast. Curr Biol 1999; 9:569-78; PMID:10359695; http://dx.doi.org/10.1016/S0960-9822(99)80260-7
- Heym RG, Zimmermann D, Edelmann FT, Israel L, Ökten Z, Kovar DR, Niessing D. In vitro reconstitution of an mRNA-transport complex reveals mechanisms of assembly and motor activation. J Cell Biol 2013; 203:971-84; PMID:24368805; http://dx.doi.org/10.1083/jcb.201302095
- Sladewski TE, Bookwalter CS, Hong MS, Trybus KM. Single-molecule reconstitution of mRNA transport by a class V myosin. Nat Struct Mol Biol 2013; 20:952-7; PMID:23812374; http://dx.doi.org/10.1038/nsmb.2614
- Dunn BD, Sakamoto T, Hong MS, Sellers JR, Takizawa PA. Myo4p is a monomeric myosin with motility uniquely adapted to transport mRNA. J Cell Biol 2007; 178:1193-206; PMID:17893244; http://dx.doi.org/10.1083/jcb.200707080
- Hodges AR, Krementsova EB, Trybus KM. She3p binds to the rod of yeast myosin V and prevents it from dimerizing, forming a single-headed motor complex. J Biol Chem 2008; 283:6906-14; PMID:18175803; http://dx.doi.org/10.1074/jbc.M708865200
- Chung S, Takizawa PA. Multiple Myo4 motors enhance ASH1 mRNA transport in Saccharomyces cerevisiae. J Cell Biol 2010; 189:755-67; PMID:20457760; http://dx.doi.org/10.1083/jcb.200912011
- Krementsova EB, Hodges AR, Bookwalter CS, Sladewski TE, Travaglia M, Sweeney HL, Trybus KM. Two single-headed myosin V motors bound to a tetrameric adapter protein form a processive complex. J Cell Biol 2011; 195:631-41; PMID:22084309; http://dx.doi.org/10.1083/jcb.201106146
- Amrute-Nayak M, Bullock SL. Single-molecule assays reveal that RNA localization signals regulate dynein-dynactin copy number on individual transcript cargoes. Nat Cell Biol 2012; 14:416-23; PMID:22366687; http://dx.doi.org/10.1038/ncb2446
- Bullock SL, Nicol A, Gross SP, Zicha D. Guidance of bidirectional motor complexes by mRNA cargoes through control of dynein number and activity. Curr Biol 2006; 16:1447-52; PMID:16860745; http://dx.doi.org/10.1016/j.cub.2006.05.055
- Soundararajan HC, Bullock SL. The influence of dynein processivity control, MAPs, and microtubule ends on directional movement of a localising mRNA. Elife (Cambridge) 2014; 3:e01596; PMID:24737859; http://dx.doi.org/10.7554/eLife.01596
- Schmid M, Jaedicke A, Du TG, Jansen R-P. Coordination of endoplasmic reticulum and mRNA localization to the yeast bud. Curr Biol 2006; 16:1538-43; PMID:16890529; http://dx.doi.org/10.1016/j.cub.2006.06.025
- Genz C, Fundakowski J, Hermesh O, Schmid M, Jansen RP. Association of the yeast RNA-binding protein She2p with the tubular endoplasmic reticulum depends on membrane curvature. J Biol Chem 2013; 288:32384-93; PMID:24056370; http://dx.doi.org/10.1074/jbc.M113.486431
- Chartrand P, Meng XH, Singer RH, Long RM. Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr Biol 1999; 9:333-6; PMID:10209102; http://dx.doi.org/10.1016/S0960-9822(99)80144-4
- Haim L, Zipor G, Aronov S, Gerst JE. A genomic integration method to visualize localization of endogenous mRNAs in living yeast. Nat Methods 2007; 4:409-12; PMID:17417645
- Trcek T, Chao JA, Larson DR, Park HY, Zenklusen D, Shenoy SM, Singer RH. Single-mRNA counting using fluorescent in situ hybridization in budding yeast. Nat Protoc 2012; 7:408-19; PMID:22301778; http://dx.doi.org/10.1038/nprot.2011.451
- Keene JD, Tenenbaum SA. Eukaryotic mRNPs may represent posttranscriptional operons. Mol Cell 2002; 9:1161-7; PMID:12086614; http://dx.doi.org/10.1016/S1097-2765(02)00559-2
- Müller M, Heuck A, Niessing D. Directional mRNA transport in eukaryotes: lessons from yeast. Cell Mol Life Sci 2007; 64:171-80; PMID:17131056; http://dx.doi.org/10.1007/s00018-006-6286-1
- Besse F, López de Quinto S, Marchand V, Trucco A, Ephrussi A. Drosophila PTB promotes formation of high-order RNP particles and represses oskar translation. Genes Dev 2009; 23:195-207; PMID:19131435; http://dx.doi.org/10.1101/gad.505709
- Michelitsch MD, Weissman JS. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc Natl Acad Sci U S A 2000; 97:11910-5; PMID:11050225; http://dx.doi.org/10.1073/pnas.97.22.11910
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 2012; 149:753-67; PMID:22579281; http://dx.doi.org/10.1016/j.cell.2012.04.017
- Kwon I, Kato M, Xiang S, Wu L, Theodoropoulos P, Mirzaei H, Han T, Xie S, Corden JL, McKnight SL. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell 2013; 155:1049-60; PMID:24267890; http://dx.doi.org/10.1016/j.cell.2013.10.033
- Schwartz JC, Wang X, Podell ER, Cech TR. RNA seeds higher-order assembly of FUS protein. Cell Rep 2013; 5:918-25; PMID:24268778; http://dx.doi.org/10.1016/j.celrep.2013.11.017
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 2003; 31:3406-15; PMID:12824337; http://dx.doi.org/10.1093/nar/gkg595
