Abstract
Localization and the associated translational control of mRNA is a well established mechanism for segregating cellular protein expression. Drosophila has been instrumental in deciphering the prevailing mechanisms of mRNA localization and regulation. This review will discuss the diverse roles of mRNA localization in the Drosophila germline, the cis-elements and cellular components regulating localization and the superimposition of translational regulatory mechanisms. Despite a history of discovery, there are still many fundamental questions regarding mRNA localization that remain unanswered. Take home messages, outstanding questions and future approaches that will likely lead to resolving these unknowns in the future are summarized at the end.
Roles for mRNA Localization
To ensure intracellular proteins act at the right time and place, cells exhibit a variety of mechanisms. Unlike in S. cerevisiae where many bud tip localizing proteins do not require localization of their transcripts, the overwhelming majority of mRNA localization in the Drosophila germline is coupled with protein function. Many animals, including Drosophila, rely on maternal transcripts to coordinate early development in processes that involve assorted post-transcriptional regulatory events.Citation1 Growing evidence suggests that post-transcriptional regulation of gene expression is more prevalent than transcriptional control.Citation2,3 Drosophila, with its heavy reliance on maternal mRNAs and its numerous experimental advantages, has emerged as a top model for addressing fundamental questions underpinning both mRNA localization and translational regulation.Citation4,5
Somatic tissues also employ mRNA localization, but show integral differences to oocytes and eggs. Many somatic tissue transcripts appear more enriched on subcellular structures than tightly localized and exhibit a greater diversity of patterns suggestive of a less concerted process.Citation6 Moreover, somatic tissue transcripts have significantly shorter half-lives than maternal mRNA which is relevant when considering localization mechanisms.Citation7 mRNA localization in somatic tissues has been well discussed in other reviews and will not be addressed further in this article.Citation1,8,9 In the developing Drosophila embryo, spatial regulation of gene expression by mRNA localization results in molecular asymmetries that coordinate early patterning events and leads to the segregation of fate determinants that are important in building the germ plasm. It is now clear that the sources of these asymmetries arise from highly orchestrated mRNA localization events that occur prior to fertilization, during oogenesis.
Two Key Roles for mRNA Localization in Early and Mid Oogenesis
Drosophila oogenesis is composed of 14 morphologically defined stages (). The oocyte develops in an egg chamber composed of somatically derived follicular epithelial cells encapsulating an interconnected set of 16 germline cells.Citation10 One of these germline cells becomes the future oocyte and the remaining 15 develop as nurse cells that provide a supporting role, producing maternal mRNAs and cytoplasmic components required for oocyte and ultimately embryonic development.Citation10 Nurse cells passively and actively exchange cytoplasm through actin-rich ring canals in early and mid-oogenesis until these supporting cells apoptose at stage 10b, initiating the late phase of oogenesis. Before the regulated demise of the nurse cells, the oocyte has already achieved two key objectives through mRNA localization: 1) events essential for establishing the anterior-posterior (A/P) and dorsal-ventral (D/V) axis have been initiated by two spatially and temporally distinct rounds of gurken (grk) mRNA translationCitation11; 2) local translation of oskar (osk) mRNA sets in motion the establishment of the future germline.Citation12-15 grk mRNA is first localized in early oogenesis (stage 2–6) to the posterior pole of the oocyte. Following translation, Grk protein, a TGF-α homolog, signals to the surrounding follicle cells causing them to adopt posterior fates.Citation16,17 These follicle cells then signal back to the oocyte resulting in a rearrangement of the oocyte microtubule (MT) cytoskeletonCitation18 and the pushing of the oocyte nucleus to the anterior margin.Citation19 The signaling with follicle cells is a pivotal moment in Drosophila development as axis patterning and germline establishment depend on the new cytoskeletal configuration.Citation16,17,20
Figure 1. Stages of Drosophila oogenesis and early embryogenesis. (A) Early oogenesis- stage 6 egg chamber. grk mRNA, synthesized in the nurse cells, localizes at the posterior of the oocyte where it is translated and signals to the overlying follicle cells. (B) Mid- oogenesis- stage 8 egg chamber. bcd, grk, and osk mRNA, synthesized in the nurse cells, localize to specific regions of the oocyte. (C) Late oogenesis -stage 10b/11 egg chamber. Nurse cells dump their cytoplasm into the oocyte while ooplasmic streaming drives a unidirectional mixing of the ooplasm. bcd and nos mRNA localize to the anterior and posterior poles, respectively. (D) Late oogenesis-stage 14 egg chamber. Oogenesis is complete and the mature egg is ready for laying, a process that can be delayed for days or even weeks. (E) Early embryogenesis-stage 4 embryo. In the syncytial blastoderm, translated bcd and nos mRNA form opposing protein gradients that pattern the A/P axis. (F) Early embryogenesis- stage 5 embryo. The blastoderm is cellularized.
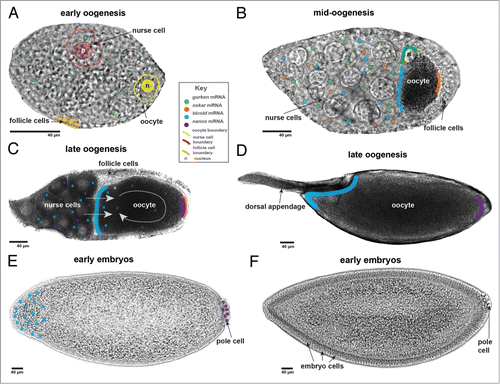
A second round of grk mRNA localization occurs at mid-oogenesis (stage 7–10a) and results in the formation of a tight RNA cap over the oocyte nucleus, which is now anteriorly located.Citation16 grk mRNA translation again results in a signaling cascade with the surrounding follicle cells, this time defining the dorsal-ventral axis.Citation16,21,22 Both rounds of grk expressions require an intact cytoskeleton, molecular motors, a cis-acting localization sequence and trans-acting factors that are described below.Citation5
Accumulation of osk mRNA at the posterior pole takes place over the course of mid-oogenesis.Citation23 Prior to being deposited in the oocyte for localization, osk mRNA undergoes a multitude of interactions in the nucleus, nurse cell cytoplasm and ooplasm essential for its localization.Citation24 These events are well described in other reviews.Citation25,26 At the posterior pole, where Grk protein first signaled, Osk protein sets the foundation on which the future germline is built.
mRNA Localization in Late Oogenesis Establishes the A/P Body Axis
The start of late oogenesis (stage 10b-14) is marked by a second major cytoskeletal re-arrangement initiated by changes in cortical actinCitation27 that result in two dynamic phenomenon: 1) rapid extrusion of nurse cell cytoplasm into the oocyte (nurse cell dumping)Citation10; 2) unidirectional churning or mixing of the ooplasm (ooplasmic streaming).Citation28 The reorganization of the ooctye MTs into the bundles at the cortex is important not only because of their requirement for ooplasmic streaming but it means that there are no longer MTs that could transport mRNAs to the posterior. During these late stages of oogenesis, nanos (nos) and bicoid (bcd) mRNA are localized, to the posterior and anterior poles respectively, by two different mechanisms.Citation29,30 At its destination, nos mRNA is translated while bcd mRNA remains translationally repressed until after fertilization.Citation31,32 Egg laying can be delayed for days or even weeks by physical or environmental strategies. Importantly, the localized mRNAs remain intact and properly positioned such that normal development can ensue once the eggs are fertilized. This raises many intriguing questions about the extended stability of mRNA in late stage oocytes.
Following the events of egg activation, fertilization and egg deposition, opposing protein gradients of Bcd and Nos form that pattern the A/P axis of the developing embryo.Citation32-35 Live imaging has shown nos mRNA actively segregates with Vasa (Vas) protein into the germ cell progenitors, the pole cells, that bud from the posterior of the embryo following fertilization.Citation36 In addition to its patterning role, nos is also required for the transcriptional and mitotic quiescence of the germ cells and their migration to the gonad. Numerous mRNAs, in addition to nos, are localized to the germ plasm and inherited by the germ cells. These transcripts presumably encode proteins that specify germ cell fate and provide a maternal supply for the early development of the germ cells, before becoming transcriptionally active.
The generation of the anterior to posterior Bcd protein gradient has been extensively studied.Citation37-39 It has become clear that bcd mRNA is not a point source, but rather both the mRNA and protein form gradients.Citation40 However, detection of single bcd mRNA particles shows that mRNA distribution alone is not sufficient to explain the observed Bcd protein gradient and it is hypothesized that active or passive protein movement is required.Citation41 There is also evidence that Grk protein can be localized in the egg chamber independently of the mRNA. In germline clones of Hephaestus, the ortholog of polypyrimidine tract-binding protein (PTB), grk mRNA localization is wild-type with Grk protein failing to be restricted to the D/A corner.Citation42
Mechanisms of mRNA Localization
Genetic screens, mutant analysis, genome-wide approaches and biochemistry experiments have all helped to establish the variety of unique and conserved trans-acting proteins and cis-acting sequences that are required for accumulating mRNAs in discrete destination. Regardless of mechanism, cis-acting elements in the mRNA are required for trans-acting proteins to recognize and bind. These interactions build a ribonucleoprotein (RNP) complex with many protein factors and mRNAs of the same or even different species. Cis-acting elements are typically in the 3′ untranslated region (UTR), where hindrance with translation is unlikely, and generally form secondary structures including stem loops, hairpins and bulges.Citation43-46 Alternative splicing of the mRNA in the nucleus can adjust these elements, resulting in unique RNP compositions and ultimately different outcomes for the mRNA.Citation47,48
The prevailing model of localization suggests that newly formed RNPs are recognized by other factors, such as proteins linking cargo to molecular motors in the case of transport, and become localized. During this localization, mRNAs are thought to be kept translationally silent. Once at their destination, RNP complexes are then remodeled and repression factors replaced by anchoring and/or translational machinery.
There are three main mechanisms of mRNA localization: active transport, diffusion-entrapment and local protection from mRNA degradation.Citation49 The ease of culturing and imaging Drosophila egg chambers and early embryos and the ability to monitor mRNA localization in both live and fixed tissue has facilitated the dissection of these mechanisms.
Active Transport in the Early Embryo
The embryo has been an especially attractive model for RNA localization due to its amenability to microinjection of fluorescently labeled mRNAs. This led to the first direct evidence of Dynein mediated mRNA transport along microtubules.Citation50 Shortly after, it was shown that the Egalitarian (Egl) - Bicaudal D (BicD) - Dynein mediates transport of mRNA to the apical side of embryo cells as well as from the nurse cells to the oocyte.Citation51 This complex has been further dissected to show that Egl, and not BicD, is capable of binding RNA sequences from many different transcripts.Citation52 A recent biochemical screen identified a novel protein, Lissencephaly-1 (Lis-1), as being required for recruiting the Dynein-Dynactin complex to localizing RNPs and, in lis-1 mutant embryos, travel distances in the minus-end direction on MTs are reduced.Citation53 High resolution tracking of mRNA using an in vitro motility assay showed processive and diffusive movements of mRNAs bound to the Dynein-Dynactin complex with mRNA localization signals increasing the processive movements.Citation54 In addition, MT associated proteins and encounters with the ends of MTs have a clear influence on RNP movement and directionality.Citation54
Whether the transport particle is moving as a single mRNA or a collection of many transcripts remains an area of debate. Single molecule fluorescence detection provides strong evidence, in vitro and in vivo, that apically localized RNAs are transported individually.Citation43 Whether this is also the case in for mRNAs in the oocyte is not clear. Future in vitro and RNA injection experiments will likely continue to lead the way for experiments testing the in vivo composition of RNP complexes and their stoichiomerty.
Active Transport in Early and Mid-Oogenesis
The predominant mechanism in early and mid-oogenesis is active transport on cytoskeletal tracks by molecular motors. In vivo labeling of endogenous grk, osk and bcd mRNA showed that each displays dynamic movements.Citation23,30,55,56 MTs impact the localization process by dictating where complexes can be transported by motors. In oogenesis, the MT cytoskeleton display three distinct arrangements, with both the population and individual MTs being highly dynamic and displaying random but biased polarity in at least some stages.Citation57 MTs in the early egg chamber are nucleated from a microtubule-organizing center at the posterior pole of the oocyte and extend anteriorly into the nurse cells.Citation58,59 grk mRNA synthesized in the nurse cells is actively transported by Dynein to the minus end of the MTs at the oocyte posterior pole.Citation16,21,60
Following the first re-arrangement of the cytoskeleton in the egg chamber, grk mRNA is actively transported in a Egl, Bic-D, and Dynein dependent manner toward the minus ends of MTs which in mid-oogenesis emanate, at least in part, from the anterior and dorsal anterior corner of the oocyte.Citation61,62 The current model, based on tracking of injected fluorescent RNA directly into the center of the oocyte, suggests two steps are involved to form the D/A cap of grk mRNA over the nucleus.Citation61 In the first step grk mRNA is actively transported to the anterior margin and in the second to the dorsal anterior corner in a step that is disrupted in squid or K10 mutants.Citation17,61 At the D/A corner, injected grk RNA enters into large non-membranous electron-dense structures that are lost when anti-Dynein heavy chain is injected.Citation62 Fluorescence recovery after photobleaching (FRAP) experiments using MS2 tagged grk mRNA shows a decrease in the mRNA's dynamics as oogenesis progresses.Citation56 In K10 and sqd mutants, grk mRNA mis-localized at the anterior is more dynamic supporting a model for these proteins in anchoring roles rather than mediating transport.Citation56 What factor or combination of factors is required for this temporal anchoring of grk mRNA still remains unclear as point mutations in the grk localization signal (GLS) suggest a least one novel factor.Citation63 Whether grk mRNA is transported with the oocyte nucleus or if it is degraded between the two localization events is unknown. A model where new grk mRNA from the adjacent nurse cells enters the oocyte at stage 7–10, is yet to be supported experimentally.
Recent genetic screens continue to identify new proteins required for mRNA localization events.Citation64,65 While some trans-factors appear to be specific to a single mRNA, many proteins are required for more than one localization event. For example, the Egl-BicD-Dynein complex is used to target mRNAs in various Drosophila cell types.Citation51,66,67 Moreover, glutathione-RNA chromatography, an efficient and specific technique for protein purification, reveals that Drosophila Syncrip, a homolog of mammalian SYNCRIP/hnRNPQ, binds grk and osk mRNA in vitro and is required for localization and translation in vivo.Citation68 Co-localization of trans-acting factors with different mRNAs that have discrete localizations raises the question of what the factors that dictates localization are.
To this end, work on osk mRNA, which sets up the future pole plasm in mid-oogenesis, has sought to address what specific factors are required for localization. Direct tagging of osk mRNA shows a bias random walk mediated by the plus-end directed motor kinesin to the posterior pole on a marginally polarized MT cytoskeleton.Citation23 Stau, mago, barentsz, and Tropomyosin II mutants result in mislocalization of osk mRNA.Citation23 At the posterior, the actin cytoskeleton, the actin binding proteins Lasp, Didum, Spire, Cappuccino, the Myosin-V motor, MTs and Dynein are all required for proper posterior accumulation and maintenance.Citation69-75 The involvement of both kinesin and Dynein in osk mRNA localization suggests that RNP particles are exposed to opposing directional forces during localization. How particles in vivo transition between plus and minus end movement remains unclear and an aim for future research. Increasingly data also suggests that localization-incompetent RNA molecules can hitch-hike on transport particles.Citation76-78 The osk 3′UTR mediates hitch-hiking, presumably through RNA-RNA interactions, however the full details of how an RNA is recognized, attached and released from a transport particle is yet to be explained.
bcd mRNA shows two recognizable phases of localization, in mid- and late oogenesis respectively. Unlike grk mRNA, which is translated and secreted in both the posterior and D/A locations, bcd mRNA is localized to the anterior where it remains translationally silent until after deposition.Citation32,33 Classic genetic analysis identified three trans-acting factors that are required for bcd localization, Exuperantia (Exu), Swallow (Swa) and Staufen (Stau).Citation79 It is clear from experiments in which bcd mRNA is first injected into a nurse cell, removed and re-injected into an ooctye that Exu, an RNA binding protein, is required in the nurse cells but not in the oocyte for anterior bcd accumulation.Citation80 This was the first evidence that established a specific role in mRNA localization for nurse cell cytoplasm, an area in the cell that is still not fully understood. Swa, which was thought to be the linking factor between bcd mRNA and Dynein,Citation81 has now been shown to act in organizing the cytoskeletal architecture that provides the microtubule tracks for bcd mRNA to move to the anterior on.Citation82 Moreover, hu li tai shao (hts), an actin regulating factor, has also been shown to play a similar roleCitation83 suggesting that proteins thought to be direct may instead be indirectly functioning in mRNA localization. The other trans-acting factor originally identified in bcd mRNA accumulation at the anterior, Stau, is not required until the second phase of localization. Interestingly, a recent genome-wide sequence analysis has shown that Stau bound transcripts have significantly longer 3′ UTRs with specific secondary structures likely to be essential for recognizing specific mRNA.Citation84
The observation that bcd and grk mRNA both require Dynein and a polarized cytoskeleton and accumulate together in stage 7 oocytes raises the question of how they ultimately adopt unique patterns. One possibility is that when grk and bcd mRNA enter the mid-stage oocyte along the anterior margin, they are simply shuttled toward the minus ends of the MTs. The overall orientation of the dynamic MTs result in grk and bcd mRNA particles moving along the entire anterior. When grk RNPs come into contact with the D/A, they become trapped in a Squid dependent manner while bcd remains mobile at the anterior. While direct evidence is lacking, grk mRNA is likely degraded after it signals to the follicle cells at the D/A corner and bcd mRNA is also likely degraded until the Stau-dependent second phase of localization is complete.
Multiple Mechanisms of Localization in Late Oogenesis
The majority of bcd mRNA is localized during late oogenesis. Stage 10b, when streaming and dumping commence, demarcates the second phase of bcd mRNA localization where it is estimated that at least 10-fold more of the mRNA accumulates than in mid-oogenesis.Citation30 Live cell imaging has shown a continual active transport mechanism requiring Dynein and a population of MTs nucleated at the anterior, before being anchored by actin at stage 14, the end of oogenesis.Citation30,55 Stau, a double stranded RNA binding protein, is essential for osk mRNA localization, translational activation and anchoringCitation12,14,Citation85-87 and is also required for bcd mRNA localization, specifically in late oogenesis.Citation30
Concurrent with bcd late-oogenesis localization, nos mRNA becomes distributed throughout the ooplasm by diffusion, facilitated by ooplasmic streaming. nos mRNA that encounters the posterior cortex of the oocyte is entrapped in association with germ plasm resident proteins, such as Vas, and anchored to the actin cytoskeleton at the posterior pole as observed by live cell imaging.Citation29 Germ cell-less and cyclin B mRNA also become localized to the posterior pole in this manner.Citation4 This mechanism is inefficient, leaving the majority (96%) of nos mRNA unlocalized. This unlocalized RNA is translationally repressed (see below) and ultimately degraded in the blastoderm embryo.Citation88 For long-term maintenance of the germ plasm, live imaging shows that Kinesin, Dynein and cortical MTs are required for germ plasm RNP, containing nos mRNA, motility which is coordinated with Myosin V mediated tethering.Citation75 This multiple mechanism model stresses the importance of restricting mRNA to the correct cellular position for extended periods of time.
Superimposition of Translational Regulatory Mechanisms
Regulating translation, both activation and repression, and degradation of mRNA is essential for cell function. At the destination, the RNP encounters or adds factors that maintain the localization and likely undergoes a large scale change in composition that results in translation. One established control mechanism is competition for interaction with the cap-binding eukaryotic initiation factor 4E (eIF4E) by eIF4G and eIF4E-binding proteins.Citation89 Once eIF4E binds to the 7-methyl guanosine cap at the 5′ end, translation can be initiated by the subsequent binding of eIF4G which, in conjunction with eIF3, leads to recruitment of the 43S ribosomal pre-initiation complex.Citation90 Unlocalized or repressed mRNAs often have eIF4E-binding proteins bound to eIF4E, thus blocking this initiation. This process is reviewed in detail.Citation89
Extensive work on osk has shown that repression is sustained through an interaction between the 5′ and 3′ ends. In this mechanism, Cup binds eIF4E via a conserved binding sequence in addition to interacting with Bruno which directly binds, with HRP48, at three sites in the 3′ UTR.Citation25,91,92 Mutant analysis of cup shows that it also negatively regulates orb, a CPEB homolog, and blocks its positive autoregulatory loop.Citation93 Regulation by cup is important as Orb protein has been shown to act as a translational activator in mid-oogenesis of both osk and grk mRNA.Citation94,95 Oligomerization of osk mRNA into large silencing particles is thought to be important in protecting transcripts from ribosomes.Citation96 Proteins are also important in this repression as shown by mutations in Polypyrimidine tract-binding (PTB) protein that decrease the size of osk particle and mis-express the mRNA.Citation97
Different mRNAs appear to be localized to the same cellular regions but experience different translational outcome. This is the case with grk and bcd mRNA in stage 8 and 9 oocytes when grk is expressed and bcd is repressed. In situ hybridization on ultra-thin frozen sections followed by immuno-electron microscopy has shown that both endogenous mRNA species associate at the anterior margin with electron dense bodies that have similar protein distributions to Processing bodies (P bodies), distinct cytoplasmic regions of mRNA control and turnover, well described in yeast.Citation98-101 bcd mRNA is detected inside the P body where translation is not supported while grk mRNA is associated at the edge where the translational activator Orb and ribosomes are present. It remains unclear if degradation follows translation of grk mRNA or if bcd mRNA is degraded at these stages as well.Citation101 At the posterior of the oocyte, P bodies are also present and their formation has been shown to be promoted in the germline by Drosophila Ge-1.Citation102 P bodies in the posterior might play a role in regulating the major differences in the translational timing of germ plasm RNAs; nos mRNA translated immediately on localization, germ cell-less translated in the early embryo, polar granule component not translated until after pole cell formation, and others not apparently translated until germ cell migration.Citation103
Take Home Messages about mRNA Localization
“When” is as critical as “where.” mRNA localization can concentrate protein expression in regions where these proteins function and, when coupled with translational control, can prevent toxicity due to production at inappropriate locations. Temporal control can be imposed on the spatial control in two ways: first, transcripts can be transported at different times—second, regardless of when transcripts are transported, they can be translated at a later time in response to a developmental or extracellular cue.
There are conserved aspects across transcripts, tissues and organisms. For example, transport complexes, such as Egl-BicD-Dynein, are used to dynamically target mRNAs in various cell typesCitation51,66,67; proteins, such as Stau, are required for mRNAs localized in the same tissue to different regions; organisms show conservation of mechanism for organizing mRNAsCitation1,8
Consider the tissue since mRNA localization is utilized for different cellular needs. In neurons the requirements are very different from the germline, with speed and readiness being essential for function.
mRNA localization and translational control has implications for human health. Alternate splicing of mRNA and changing RNP complex composition enable cells to alter and tune their transcriptome and proteome in response to internal and external cues. The increasing identification of mutations in mRNA that cause disease has lead to therapeutics targeting mRNAs and RNPs.Citation104
Outstanding questions about mRNA localization
To what extent are mRNPs heterogeneous and are these complexes in a state of continued flux? Are mRNA complexes remodelled after nuclear export or are they fully equipped when they set out into the cytoplasm? Does the mRNP shed components after a function is complete? How dynamic are these complexes?
Is there a quality control check point in the formation or localization of mRNPs? Is the complex destroyed or rebuilt if the wrong composition is detected?
In a cell with multiple populations of MTs, how do mRNAs discriminate between these populations? Is there a general mechanism organizing the availability and location of transport components?
What is the complete mechanism of how mRNAs are translationally activated? Are mRNAs translated within the RNPs or do they have to be released from the RNP?
To what extent does mRNA act as a scaffold for protein-protein interactions or miRNA-protein interactions? Are there localized mRNAs that play a structural role rather than serving as templates for protein synthesis?
Is miRNA-based regulation superimposed on mRNA localization? What is the full extent and mechanism of miRNA function with localized transcripts? Are there widespread requirements for miRNA beyond clearance of mRNAs.Citation105-108
Future Experimental Approaches
Ongoing proteomic analysis, high throughput biochemical approaches and advanced imaging methods are all working toward addressing these and many more key questions about mRNA localization. The understanding of cellular mechanism will require not only these techniques, but also in vivo experimentation.
Use of single molecule analysis is increasing and experiments that follow an individual mRNA through a living tissue while simultaneously visualizing key components in real-time are a realistic goal. Experiments such as this will have to address the formation, remodeling and degradation of RNPs in vivo.
System-wide identification of RNA binding proteinsCitation109 and genome-wide predictions of protein binding sites provide an insight into an enormous number of testable in vivo hypotheses. Future iterations of the interactome of substantial interest will be in defining the ratios of protein-RNA interactions in vivo. Sampling mRNA temporally and spatially and ascertaining complexes stoichiometry in vivo will offer key insights for addressing the dynamics of mRNA associations.Citation110
CRISPR/Cas genome editing technology promises efficient germline and somatic engineering in Drosophila.Citation111 For example, the manipulation of mRNA protein binding sites in vivo will be simplified and made significantly more time efficient. As this technology becomes fully implemented and further advanced, new experimental approaches will be applied.
Fast temporal control over protein function. Current techniques such as photo inactivation by light including chromophore assisted light inactivation (CALI),Citation112,113 transgenically Encoded Protein Photoinactivation (FIAsH-FALI)Citation114 and Channelrhodopsin-2 (ChR2)Citation115 as well as the tobacco etch mosaic virus (TEV) proteaseCitation116 and auxinCitation117 based methods have not been widely adapted to questions of mRNA localization. In part, the specificity and speed of these techniques render them incompatible. Advancements or new methods of switching off a protein in vivo will be important in further understanding questions addressing when and where proteins function.
The Drosophila germline tissue is an ideal model for implementing these and other new approaches that will ultimately result in a clear understanding of this common and essential cellular process.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
I am grateful to Elizabeth R. Gavis for discussions and critical reading of the manuscript. Also to Alexander Davidson for feedback on drafts.
Funding
This work was supported by the University of Cambridge, ISSF to T.T.W. [grant number 097814].
References
- Medioni C, Mowry K, Besse F. Principles and roles of mRNA localization in animal development. Development 2012; 139:3263-76; PMID:22912410; http://dx.doi.org/10.1242/dev.078626
- Weill L, Belloc E, Bava F-A, Méndez R. Translational control by changes in poly(A) tail length: recycling mRNAs. Nat Struct Mol Biol 2012; 19:577-85; PMID:22664985; http://dx.doi.org/10.1038/nsmb.2311
- Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Mol Cell 2011; 43:853-66; PMID:21925375; http://dx.doi.org/10.1016/j.molcel.2011.08.017
- Becalska AN, Gavis ER. Lighting up mRNA localization in Drosophila oogenesis. Development 2009; 136:2493-503; PMID:19592573; http://dx.doi.org/10.1242/dev.032391
- Lasko P. mRNA localization and translational control in Drosophila oogenesis. Cold Spring Harb Perspect Biol 2012; 4:4; PMID:22865893; http://dx.doi.org/10.1101/cshperspect.a012294
- Lécuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 2007; 131:174-87; PMID:17923096; http://dx.doi.org/10.1016/j.cell.2007.08.003
- Thomsen S, Anders S, Janga SC, Huber W, Alonso CR. Genome-wide analysis of mRNA decay patterns during early Drosophila development. Genome Biol 2010; 11:R93; PMID:20858238; http://dx.doi.org/10.1186/gb-2010-11-9-r93
- Holt CE, Bullock SL. Subcellular mRNA localization in animal cells and why it matters. Science 2009; 326:1212-6; PMID:19965463; http://dx.doi.org/10.1126/science.1176488
- Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell 2009; 136:719-30; PMID:19239891; http://dx.doi.org/10.1016/j.cell.2009.01.04410
- Spradling AC. Developmental genetics of oogenesis. In The Development of Drosophila melanogaster, M. Bate and A. Martinez Arias, eds. Cold Spring Harbor, NY: Cold Spring Harbor Press 1993; pp.1-70.
- Nilson LA, Schüpbach T. EGF receptor signaling in Drosophila oogenesis. Curr Top Dev Biol 1999; 44:203-43; PMID:9891881; http://dx.doi.org/10.1016/S0070-2153(08)60471-8
- Ephrussi A, Dickinson LK, Lehmann R. Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell 1991; 66:37-50; PMID:2070417; http://dx.doi.org/10.1016/0092-8674(91)90137-N
- Lehmann R, Nüsslein-Volhard C. Abdominal segmentation, pole cell formation, and embryonic polarity require the localized activity of oskar, a maternal gene in Drosophila. Cell 1986; 47:141-52; PMID:3093084; http://dx.doi.org/10.1016/0092-8674(86)90375-2
- Kim-Ha J, Smith JL, Macdonald PM. oskar mRNA is localized to the posterior pole of the Drosophila oocyte. Cell 1991; 66:23-35; PMID:2070416; http://dx.doi.org/10.1016/0092-8674(91)90136-M
- Ephrussi A, Lehmann R. Induction of germ cell formation by oskar. Nature 1992; 358:387-92; PMID:1641021; http://dx.doi.org/10.1038/358387a0
- González-Reyes A, Elliott H, St Johnston D. Polarization of both major body axes in Drosophila by gurken-torpedo signalling. Nature 1995; 375:654-8; PMID:7791898; http://dx.doi.org/10.1038/375654a0
- Neuman-Silberberg FS, Schüpbach T. The Drosophila dorsoventral patterning gene gurken produces a dorsally localized RNA and encodes a TGF alpha-like protein. Cell 1993; 75:165-74; PMID:7691414; http://dx.doi.org/10.1016/0092-8674(93)90688-M
- González-Reyes A, St Johnston D. Patterning of the follicle cell epithelium along the anterior-posterior axis during Drosophila oogenesis. Development 1998; 125:2837-46; PMID:9655806
- Zhao T, Graham OS, Raposo A, St Johnston D. Growing microtubules push the oocyte nucleus to polarize the Drosophila dorsal-ventral axis. Science 2012; 336:999-1003; PMID:22499806; http://dx.doi.org/10.1126/science.1219147
- Steinhauer J, Kalderon D. Microtubule polarity and axis formation in the Drosophila oocyte. Dev Dyn 2006; 235:1455-68; PMID:16586443; http://dx.doi.org/10.1002/dvdy.20770
- Roth S, Neuman-Silberberg FS, Barcelo G, Schüpbach T. cornichon and the EGF receptor signaling process are necessary for both anterior-posterior and dorsal-ventral pattern formation in Drosophila. Cell 1995; 81:967-78; PMID:7540118; http://dx.doi.org/10.1016/0092-8674(95)90016-0
- Neuman-Silberberg FS, Schüpbach T. Dorsoventral axis formation in Drosophila depends on the correct dosage of the gene gurken. Development 1994; 120:2457-63; PMID:7956825
- Zimyanin VL, Belaya K, Pecreaux J, Gilchrist MJ, Clark A, Davis I, St Johnston D. In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell 2008; 134:843-53; PMID:18775316; http://dx.doi.org/10.1016/j.cell.2008.06.053
- Ghosh S, Marchand V, Gáspár I, Ephrussi A. Control of RNP motility and localization by a splicing-dependent structure in oskar mRNA. Nat Struct Mol Biol 2012; 19:441-9; PMID:22426546; http://dx.doi.org/10.1038/nsmb.2257
- Kato Y, Nakamura A. Roles of cytoplasmic RNP granules in intracellular RNA localization and translational control in the Drosophila oocyte. Dev Growth Differ 2012; 54:19-31; PMID:22111938; http://dx.doi.org/10.1111/j.1440-169X.2011.01314.x
- Marchand V, Gaspar I, Ephrussi A. An intracellular transmission control protocol: assembly and transport of ribonucleoprotein complexes. Curr Opin Cell Biol 2012; 24:202-10; PMID:22278045; http://dx.doi.org/10.1016/j.ceb.2011.12.014
- Dahlgaard K, Raposo AASF, Niccoli T, St Johnston D. Capu and Spire assemble a cytoplasmic actin mesh that maintains microtubule organization in the Drosophila oocyte. Dev Cell 2007; 13:539-53; PMID:17925229; http://dx.doi.org/10.1016/j.devcel.2007.09.003
- Gutzeit HO. The role of microfilaments in cytoplasmic streaming in Drosophila follicles. J Cell Sci 1986; 80:159-69; PMID:3722280
- Forrest KM, Gavis ER. Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr Biol 2003; 13:1159-68; PMID:12867026; http://dx.doi.org/10.1016/S0960-9822(03)00451-2
- Weil TT, Forrest KM, Gavis ER. Localization of bicoid mRNA in late oocytes is maintained by continual active transport. Dev Cell 2006; 11:251-62; PMID:16890164; http://dx.doi.org/10.1016/j.devcel.2006.06.006
- Forrest KM, Clark IE, Jain RA, Gavis ER. Temporal complexity within a translational control element in the nanos mRNA. Development 2004; 131:5849-57; PMID:15525666; http://dx.doi.org/10.1242/dev.01460
- Driever W, Nüsslein-Volhard C. A gradient of bicoid protein in Drosophila embryos. Cell 1988; 54:83-93; PMID:3383244; http://dx.doi.org/10.1016/0092-8674(88)90182-1
- Driever W, Nüsslein-Volhard C. The bicoid protein determines position in the Drosophila embryo in a concentration-dependent manner. Cell 1988; 54:95-104; PMID:3383245; http://dx.doi.org/10.1016/0092-8674(88)90183-3
- Gavis ER, Lehmann R. Localization of nanos RNA controls embryonic polarity. Cell 1992; 71:301-13; PMID:1423595; http://dx.doi.org/10.1016/0092-8674(92)90358-J
- Wang C, Lehmann R. Nanos is the localized posterior determinant in Drosophila. Cell 1991; 66:637-47; PMID:1908748; http://dx.doi.org/10.1016/0092-8674(91)90110-K
- Lerit DA, Gavis ER. Transport of germ plasm on astral microtubules directs germ cell development in Drosophila. Curr Biol 2011; 21:439-48; PMID:21376599; http://dx.doi.org/10.1016/j.cub.2011.01.073
- Gregor T, Wieschaus EF, McGregor AP, Bialek W, Tank DW. Stability and nuclear dynamics of the bicoid morphogen gradient. Cell 2007; 130:141-52; PMID:17632061; http://dx.doi.org/10.1016/j.cell.2007.05.026
- Gregor T, Tank DW, Wieschaus EF, Bialek W. Probing the limits to positional information. Cell 2007; 130:153-64; PMID:17632062; http://dx.doi.org/10.1016/j.cell.2007.05.025
- Lipshitz HD. Follow the mRNA: a new model for Bicoid gradient formation. Nat Rev Mol Cell Biol 2009; 10:509-12; PMID:19626044; http://dx.doi.org/10.1038/nrm2730
- Spirov A, Fahmy K, Schneider M, Frei E, Noll M, Baumgartner S. Formation of the bicoid morphogen gradient: an mRNA gradient dictates the protein gradient. Development 2009; 136:605-14; PMID:19168676; http://dx.doi.org/10.1242/dev.031195
- Little SC, Tkačik G, Kneeland TB, Wieschaus EF, Gregor T. The formation of the Bicoid morphogen gradient requires protein movement from anteriorly localized mRNA. PLoS Biol 2011; 9:e1000596; PMID:21390295; http://dx.doi.org/10.1371/journal.pbio.1000596
- McDermott SM, Davis I. Drosophila Hephaestus/polypyrimidine tract binding protein is required for dorso-ventral patterning and regulation of signalling between the germline and soma. PLoS One 2013; 8:e69978; PMID:23894566; http://dx.doi.org/10.1371/journal.pone.0069978
- Amrute-Nayak M, Bullock SL. Single-molecule assays reveal that RNA localization signals regulate dynein-dynactin copy number on individual transcript cargoes. Nat Cell Biol 2012; 14:416-23; PMID:22366687; http://dx.doi.org/10.1038/ncb2446
- Hamilton RS, Davis I, Medioni C, Davis I, Lin S, Mowry K, Zhang S, Besse F, Cohen RS. RNA localization signals: deciphering the message with bioinformatics. Semin Cell Dev Biol 2007; 18:178-85; PMID:17452113; http://dx.doi.org/10.1016/j.semcdb.2007.02.001
- Patel VL, Mitra S, Harris R, Buxbaum AR, Lionnet T, Brenowitz M, Girvin M, Levy M, Almo SC, Singer RH, et al. Spatial arrangement of an RNA zipcode identifies mRNAs under post-transcriptional control. Genes Dev 2012; 26:43-53; PMID:22215810; http://dx.doi.org/10.1101/gad.177428.111
- Wilhelm JE, Vale RD. RNA on the move: the mRNA localization pathway. J Cell Biol 1993; 123:269-74; PMID:8408211; http://dx.doi.org/10.1083/jcb.123.2.269
- Horne-Badovinac S, Bilder D. Dynein regulates epithelial polarity and the apical localization of stardust A mRNA. PLoS Genet 2008; 4:e8; PMID:18208331; http://dx.doi.org/10.1371/journal.pgen.0040008
- Trcek T, Singer RH. The cytoplasmic fate of an mRNP is determined cotranscriptionally: exception or rule? Genes Dev 2010; 24:1827-31; PMID:20810644; http://dx.doi.org/10.1101/gad.1972810
- Meignin C, Davis I. Transmitting the message: intracellular mRNA localization. Curr Opin Cell Biol 2010; 22:112-9; PMID:20022233; http://dx.doi.org/10.1016/j.ceb.2009.11.011
- Wilkie GS, Davis I. Drosophila wingless and pair-rule transcripts localize apically by dynein-mediated transport of RNA particles. Cell 2001; 105:209-19; PMID:11336671; http://dx.doi.org/10.1016/S0092-8674(01)00312-9
- Bullock SL, Ish-Horowicz D. Conserved signals and machinery for RNA transport in Drosophila oogenesis and embryogenesis. Nature 2001; 414:611-6; PMID:11740552; http://dx.doi.org/10.1038/414611a
- Dienstbier M, Boehl F, Li X, Bullock SL. Egalitarian is a selective RNA-binding protein linking mRNA localization signals to the dynein motor. Genes Dev 2009; 23:1546-58; PMID:19515976; http://dx.doi.org/10.1101/gad.531009
- Dix CI, Soundararajan HC, Dzhindzhev NS, Begum F, Suter B, Ohkura H, Stephens E, Bullock SL. Lissencephaly-1 promotes the recruitment of dynein and dynactin to transported mRNAs. J Cell Biol 2013; 202:479-94; PMID:23918939; http://dx.doi.org/10.1083/jcb.201211052
- Soundararajan HC, Bullock SL. The influence of dynein processivity control, MAPs, and microtubule ends on directional movement of a localising mRNA. Elife 2014; 3:e01596; PMID:24737859; http://dx.doi.org/10.7554/eLife.01596
- Weil TT, Parton R, Davis I, Gavis ER. Changes in bicoid mRNA anchoring highlight conserved mechanisms during the oocyte-to-embryo transition. Curr Biol 2008; 18:1055-61; PMID:18639459; http://dx.doi.org/10.1016/j.cub.2008.06.046
- Jaramillo AM, Weil TT, Goodhouse J, Gavis ER, Schupbach T. The dynamics of fluorescently labeled endogenous gurken mRNA in Drosophila. J Cell Sci 2008; 121:887-94; PMID:18303053; http://dx.doi.org/10.1242/jcs.019091
- Parton RM, Hamilton RS, Ball G, Yang L, Cullen CF, Lu W, Ohkura H, Davis I. A PAR-1-dependent orientation gradient of dynamic microtubules directs posterior cargo transport in the Drosophila oocyte. J Cell Biol 2011; 194:121-35; PMID:21746854; http://dx.doi.org/10.1083/jcb.201103160
- Clark I, Giniger E, Ruohola-Baker H, Jan LY, Jan YN. Transient posterior localization of a kinesin fusion protein reflects anteroposterior polarity of the Drosophila oocyte. Curr Biol 1994; 4:289-300; PMID:7922338; http://dx.doi.org/10.1016/S0960-9822(00)00068-3
- Theurkauf WE, Smiley S, Wong ML, Alberts BM. Reorganization of the cytoskeleton during Drosophila oogenesis: implications for axis specification and intercellular transport. Development 1992; 115:923-36; PMID:1451668
- Januschke J, Gervais L, Gillet L, Keryer G, Bornens M, Guichet A. The centrosome-nucleus complex and microtubule organization in the Drosophila oocyte. Development 2006; 133:129-39; PMID:16319114; http://dx.doi.org/10.1242/dev.02179
- MacDougall N, Clark A, MacDougall E, Davis I. Drosophila gurken (TGFalpha) mRNA localizes as particles that move within the oocyte in two dynein-dependent steps. Dev Cell 2003; 4:307-19; PMID:12636913; http://dx.doi.org/10.1016/S1534-5807(03)00058-3
- Delanoue R, Herpers B, Soetaert J, Davis I, Rabouille C. Drosophila Squid/hnRNP helps Dynein switch from a gurken mRNA transport motor to an ultrastructural static anchor in sponge bodies. Dev Cell 2007; 13:523-38; PMID:17925228; http://dx.doi.org/10.1016/j.devcel.2007.08.022
- Lan L, Lin S, Zhang S, Cohen RS. Evidence for a transport-trap mode of Drosophila melanogaster gurken mRNA localization. PLoS One 2010; 5:e15448; PMID:21103393; http://dx.doi.org/10.1371/journal.pone.0015448
- Chang CW, Nashchekin D, Wheatley L, Irion U, Dahlgaard K, Montague TG, Hall J, St Johnston D. Anterior-posterior axis specification in Drosophila oocytes: identification of novel bicoid and oskar mRNA localization factors. Genetics 2011; 188:883-96; PMID:21625003; http://dx.doi.org/10.1534/genetics.111.129312
- Hayashi R, Wainwright SM, Liddell SJ, Pinchin SM, Horswell S, Ish-Horowicz D. A genetic screen based on in vivo RNA imaging reveals centrosome-independent mechanisms for localizing gurken transcripts in Drosophila. G3 (Bethesda) 2014; 4:749-60; PMID:24531791; http://dx.doi.org/10.1534/g3.114.010462
- Hughes JR, Bullock SL, Ish-Horowicz D. Inscuteable mRNA localization is dynein-dependent and regulates apicobasal polarity and spindle length in Drosophila neuroblasts. Curr Biol 2004; 14:1950-6; PMID:15530398; http://dx.doi.org/10.1016/j.cub.2004.10.022
- Van de Bor V, Zimniak G, Cérézo D, Schaub S, Noselli S. Asymmetric localisation of cytokine mRNA is essential for JAK/STAT activation during cell invasiveness. Development 2011; 138:1383-93; PMID:21350010; http://dx.doi.org/10.1242/dev.056184
- McDermott SM, Meignin C, Rappsilber J, Davis I. Drosophila Syncrip binds the gurken mRNA localisation signal and regulates localised transcripts during axis specification. Biol Open 2012; 1:488-97; PMID:23213441; http://dx.doi.org/10.1242/bio.2012885
- Krauss J, López de Quinto S, Nüsslein-Volhard C, Ephrussi A. Myosin-V regulates oskar mRNA localization in the Drosophila oocyte. Curr Biol 2009; 19:1058-63; PMID:19481457; http://dx.doi.org/10.1016/j.cub.2009.04.062
- Suyama R, Jenny A, Curado S, Pellis-van Berkel W, Ephrussi A. The actin-binding protein Lasp promotes Oskar accumulation at the posterior pole of the Drosophila embryo. Development 2009; 136:95-105; PMID:19036801; http://dx.doi.org/10.1242/dev.027698
- Tanaka T, Kato Y, Matsuda K, Hanyu-Nakamura K, Nakamura A. Drosophila Mon2 couples Oskar-induced endocytosis with actin remodeling for cortical anchorage of the germ plasm. Development 2011; 138:2523-32; PMID:21610029; http://dx.doi.org/10.1242/dev.062208
- Tanaka T, Nakamura A. Oskar-induced endocytic activation and actin remodeling for anchorage of the Drosophila germ plasm. Bioarchitecture 2011; 1:122-6; PMID:21922042; http://dx.doi.org/10.4161/bioa.1.3.17313
- Sanghavi P, Lu S, Gonsalvez GB. A functional link between localized Oskar, dynamic microtubules, and endocytosis. Dev Biol 2012; 367:66-77; PMID:22561189; http://dx.doi.org/10.1016/j.ydbio.2012.04.024
- Sanghavi P, Laxani S, Li X, Bullock SL, Gonsalvez GB. Dynein associates with oskar mRNPs and is required for their efficient net plus-end localization in Drosophila oocytes. PLoS One 2013; 8:e80605; PMID:24244700; http://dx.doi.org/10.1371/journal.pone.0080605
- Sinsimer KS, Lee JJ, Thiberge SY, Gavis ER. Germ plasm anchoring is a dynamic state that requires persistent trafficking. Cell Rep 2013; 5:1169-77; PMID:24290763; http://dx.doi.org/10.1016/j.celrep.2013.10.045
- Hartswood E, Brodie J, Vendra G, Davis I, Finnegan DJ. RNA:RNA interaction can enhance RNA localization in Drosophila oocytes. RNA 2012; 18:729-37; PMID:22345148; http://dx.doi.org/10.1261/rna.026674.111
- Jambor H, Brunel C, Ephrussi A. Dimerization of oskar 3′ UTRs promotes hitchhiking for RNA localization in the Drosophila oocyte. RNA 2011; 17:2049-57; PMID:22028360; http://dx.doi.org/10.1261/rna.2686411
- Jambor H, Mueller S, Bullock SL, Ephrussi A. A stem-loop structure directs oskar mRNA to microtubule minus ends. RNA 2014; 20:429-39; PMID:24572808; http://dx.doi.org/10.1261/rna.041566.113
- St Johnston D, Driever W, Berleth T, Richstein S, Nüsslein-Volhard C. Multiple steps in the localization of bicoid RNA to the anterior pole of the Drosophila oocyte. Development 1989; 107(Suppl):13-9; PMID:2483989
- Cha BJ, Koppetsch BS, Theurkauf WE. In vivo analysis of Drosophila bicoid mRNA localization reveals a novel microtubule-dependent axis specification pathway. Cell 2001; 106:35-46; PMID:11461700; http://dx.doi.org/10.1016/S0092-8674(01)00419-6
- Schnorrer F, Bohmann K, Nüsslein-Volhard C. The molecular motor dynein is involved in targeting swallow and bicoid RNA to the anterior pole of Drosophila oocytes. Nat Cell Biol 2000; 2:185-90; PMID:10783235; http://dx.doi.org/10.1038/35008601
- Weil TT, Xanthakis D, Parton R, Dobbie I, Rabouille C, Gavis ER, Davis I. Distinguishing direct from indirect roles for bicoid mRNA localization factors. Development 2010; 137:169-76; PMID:20023172; http://dx.doi.org/10.1242/dev.044867
- Pokrywka NJ, Zhang H, Raley-Susman K. Distinct roles for hu li tai shao and swallow in cytoskeletal organization during Drosophila oogenesis. Dev Dyn 2014; 243:906-16; PMID:24677508; http://dx.doi.org/10.1002/dvdy.24132
- Laver JD, Li X, Ancevicius K, Westwood JT, Smibert CA, Morris QD, Lipshitz HD. Genome-wide analysis of Staufen-associated mRNAs identifies secondary structures that confer target specificity. Nucleic Acids Res 2013; 41:9438-60; PMID:23945942; http://dx.doi.org/10.1093/nar/gkt702
- St Johnston D, Beuchle D, Nüsslein-Volhard C. Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell 1991; 66:51-63; PMID:1712672; http://dx.doi.org/10.1016/0092-8674(91)90138-O
- van Eeden FJ, Palacios IM, Petronczki M, Weston MJ, St Johnston D. Barentsz is essential for the posterior localization of oskar mRNA and colocalizes with it to the posterior pole. J Cell Biol 2001; 154:511-23; PMID:11481346; http://dx.doi.org/10.1083/jcb.200105056
- Micklem DR, Adams J, Grünert S, St Johnston D. Distinct roles of two conserved Staufen domains in oskar mRNA localization and translation. EMBO J 2000; 19:1366-77; PMID:10716936; http://dx.doi.org/10.1093/emboj/19.6.1366
- Bergsten SE, Gavis ER. Role for mRNA localization in translational activation but not spatial restriction of nanos RNA. Development 1999; 126:659-69; PMID:9895314
- Jackson RJ, Hellen CUT, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 2010; 11:113-27; PMID:20094052; http://dx.doi.org/10.1038/nrm2838
- Parton RM, Davidson A, Davis I, Weil TT. Subcellular mRNA localisation at a glance. J Cell Sci 2014; 127:2127-33; PMID:24833669; http://dx.doi.org/10.1242/jcs.114272
- Wilhelm JE, Hilton M, Amos Q, Henzel WJ. Cup is an eIF4E binding protein required for both the translational repression of oskar and the recruitment of Barentsz. J Cell Biol 2003; 163:1197-204; PMID:14691132; http://dx.doi.org/10.1083/jcb.200309088
- Nakamura A, Sato K, Hanyu-Nakamura K. Drosophila cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Dev Cell 2004; 6:69-78; PMID:14723848; http://dx.doi.org/10.1016/S1534-5807(03)00400-3
- Wong LC, Schedl P. Cup blocks the precocious activation of the orb autoregulatory loop. PLoS One 2011; 6:e28261; PMID:22164257; http://dx.doi.org/10.1371/journal.pone.0028261
- Chang JS, Tan L, Schedl P. The Drosophila CPEB homolog, orb, is required for oskar protein expression in oocytes. Dev Biol 1999; 215:91-106; PMID:10525352; http://dx.doi.org/10.1006/dbio.1999.9444
- Chang JS, Tan L, Wolf MR, Schedl P. Functioning of the Drosophila orb gene in gurken mRNA localization and translation. Development 2001; 128:3169-77; PMID:11688565
- Chekulaeva M, Hentze MW, Ephrussi A. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell 2006; 124:521-33; PMID:16469699; http://dx.doi.org/10.1016/j.cell.2006.01.031
- Besse F, Ephrussi A. Translational control of localized mRNAs: restricting protein synthesis in space and time. Nat Rev Mol Cell Biol 2008; 9:971-80; PMID:19023284; http://dx.doi.org/10.1038/nrm2548
- Decker CJ, Parker R. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol 2012; 4:a012286; PMID:22763747; http://dx.doi.org/10.1101/cshperspect.a012286
- Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 2005; 11:371-82; PMID:15703442; http://dx.doi.org/10.1261/rna.7258505
- Balagopal V, Parker R. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr Opin Cell Biol 2009; 21:403-8; PMID:19394210; http://dx.doi.org/10.1016/j.ceb.2009.03.005
- Weil TT, Parton RM, Herpers B, Soetaert J, Veenendaal T, Xanthakis D, Dobbie IM, Halstead JM, Hayashi R, Rabouille C, et al. Drosophila patterning is established by differential association of mRNAs with P bodies. Nat Cell Biol 2012; 14:1305-13; PMID:23178881; http://dx.doi.org/10.1038/ncb2627
- Fan S-J, Marchand V, Ephrussi A. Drosophila Ge-1 promotes P body formation and oskar mRNA localization. PLoS One 2011; 6:e20612; PMID:21655181; http://dx.doi.org/10.1371/journal.pone.0020612
- Rangan P, DeGennaro M, Jaime-Bustamante K, Coux R-X, Martinho RG, Lehmann R. Temporal and spatial control of germ-plasm RNAs. Curr Biol 2009; 19:72-7; PMID:19110432; http://dx.doi.org/10.1016/j.cub.2008.11.066
- Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell 2009; 136:777-93; PMID:19239895; http://dx.doi.org/10.1016/j.cell.2009.02.011
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 2006; 312:75-9; PMID:16484454; http://dx.doi.org/10.1126/science.1122689
- Bushati N, Stark A, Brennecke J, Cohen SM. Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr Biol 2008; 18:501-6; PMID:18394895; http://dx.doi.org/10.1016/j.cub.2008.02.081
- Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development 2009; 136:3033-42; PMID:19700615; http://dx.doi.org/10.1242/dev.033183
- Koebernick K, Loeber J, Arthur PK, Tarbashevich K, Pieler T. Elr-type proteins protect Xenopus Dead end mRNA from miR-18-mediated clearance in the soma. Proc Natl Acad Sci U S A 2010; 107:16148-53; PMID:20805475; http://dx.doi.org/10.1073/pnas.1004401107
- Castello A, Horos R, Strein C, Fischer B, Eichelbaum K, Steinmetz LM, Krijgsveld J, Hentze MW. System-wide identification of RNA-binding proteins by interactome capture. Nat Protoc 2013; 8:491-500; PMID:23411631; http://dx.doi.org/10.1038/nprot.2013.020
- Müller-McNicoll M, Neugebauer KM. How cells get the message: dynamic assembly and function of mRNA-protein complexes. Nat Rev Genet 2013; 14:275-87; PMID:23478349; http://dx.doi.org/10.1038/nrg3434
- Port F, Chen HM, Lee T, Bullock SL. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci U S A 2014; 111:E2967-76; PMID:25002478; http://dx.doi.org/10.1073/pnas.1405500111
- Bulina ME, Lukyanov KA, Britanova OV, Onichtchouk D, Lukyanov S, Chudakov DM. Chromophore-assisted light inactivation (CALI) using the phototoxic fluorescent protein KillerRed. Nat Protoc 2006; 1:947-53; PMID:17406328; http://dx.doi.org/10.1038/nprot.2006.89
- Vegh RB, Solntsev KM, Kuimova MK, Cho S, Liang Y, Loo BLW, Tolbert LM, Bommarius AS. Reactive oxygen species in photochemistry of the red fluorescent protein “Killer Red”. Chem Commun (Camb) 2011; 47:4887-9; PMID:21359336; http://dx.doi.org/10.1039/c0cc05713d
- Marek KW, Davis GW. Transgenically encoded protein photoinactivation (FlAsH-FALI): acute inactivation of synaptotagmin I. Neuron 2002; 36:805-13; PMID:12467585; http://dx.doi.org/10.1016/S0896-6273(02)01068-1
- Zimmermann G, Wang L-P, Vaughan AG, Manoli DS, Zhang F, Deisseroth K, Baker BS, Scott MP. Manipulation of an innate escape response in Drosophila: photoexcitation of acj6 neurons induces the escape response. PLoS One 2009; 4:e5100; PMID:19340304; http://dx.doi.org/10.1371/journal.pone.0005100
- Pauli A, Althoff F, Oliveira RA, Heidmann S, Schuldiner O, Lehner CF, Dickson BJ, Nasmyth K. Cell-type-specific TEV protease cleavage reveals cohesin functions in Drosophila neurons. Dev Cell 2008; 14:239-51; PMID:18267092; http://dx.doi.org/10.1016/j.devcel.2007.12.009
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods 2009; 6:917-22; PMID:19915560; http://dx.doi.org/10.1038/nmeth.1401
