Abstract
Long terminal repeat (LTR) retrotransposons are not only the ancient predecessors of retroviruses, but they constitute significant fractions of the genomes of many eukaryotic species. Studies of their structure and function are motivated by opportunities to gain insight into common functions of retroviruses and retrotransposons, diverse mechanisms of intracellular genomic mobility, and host factors that diminish or enhance retrotransposition. This review focuses on the nucleocapsid (NC) protein of a Saccharomyces cerevisiae LTR retrotransposon, the metavirus, Ty3. Retrovirus NC promotes genomic (g)RNA dimerization and packaging, tRNA primer annealing, reverse transcription strand transfers, and host protein interactions with gRNA. Studies of Ty3 NC have revealed key roles for Ty3 NC in formation of retroelement assembly sites (retrosomes), and in chaperoning primer tRNA to both dimerize and circularize Ty3 gRNA. We speculate that Ty3 NC, together with P-body and stress-granule proteins, plays a role in transitioning Ty3 RNA from translation template to gRNA, and that interactions between the acidic spacer domain of Ty3 Gag3 and the adjacent basic NC domain control condensation of the virus-like particle.
Introduction
Long terminal repeat (LTR) retrotransposons are ancient intracellular precursors of retroviruses.Citation1–Citation3 These elements have common functions with retrovirusesCitation4,Citation5 in that they assemble polyprotein precursors around genomic (g)RNA to form virus-like particles (VLPs), reverse transcribe the gRNA, and integrate the complementary (c)DNA copy into host chromosomes. Similar to retroviruses, a RNA-binding domain resides within the retrotransposon major structural polyprotein precursor or Gag protein and cis-acting packaging sequences (psi) reside within the RNA. For a subset of LTR retrotransposons, the processed form of the RNA-binding domain is homologous to retrovirus nucleocapsid (NC). Retrovirus NC proteins range in MW from 7 to 15 kDa. They provide aggregation and destabilization chaperone activities to allow duplex formation and annealing reactions critical for particle formation and replication. Despite its small size, NC not only performs these functions, but more recently has been implicated in aspects of VLP assembly involving interactions with a diverse array of host factors.
Retroviruses differ from retrotransposons in that they traffic to the cell surface, require envelopment for maturation, and enter new host cells prior to uncoating, reverse transcription and integration. In contrast, retrotransposons undergo maturation, reverse transcription, uncoating and integration within a single host cell. Enveloped, immature spherical retrovirus particles undergo reorganization as a result of proteolytic maturation. In contrast, retrotransposons must achieve a similar state competent to uncoat in the absence of the enveloped microenvironment. There is a considerable amount known about retrovirus NC (reviewed in ref. Citation6–Citation10); on the other hand, relatively few retrotransposon NC proteins have been investigated and much less is known. In this review we compare functions of retrovirus NC to functions of a retrotransposon NC, in order to identify common and therefore central functions, as well as different and potentially more specialized functions. The Saccharomyces cerevisiae Metaviridae Ty3 NC protein has been investigated in vivo and in vitro. In this review we focus primarily on Ty3 NC and compare its functions to the functions of retrovirus NC proteins, particularly those of human immunodeficiency virus (HIV)-1 NC. The genomics structures of Ty3 and HIV-1 are shown in .
Overview of Retrovirus and LTR Retrotransposon Structure
Retrovirus genome and protein structure.
The integrated form of the retrovirus has LTRs (U3-R-U5) flanking the coding region ().Citation4 Viral gRNA is initiated in the upstream LTR and terminated at a position downstream of the initiation site in the downstream LTR. Thus, the transcript can be described as R-U5-coding sequence-U3-R, where U5 and U3 represent sequences uniquely present on the 5′ and 3′ ends of the RNA, respectively, and R represents sequence repeated on both ends. The retrovirus genome is diploid and the two gRNAs dimerize over a several hundred-nt region to template production of a full-length cDNA copy (reviewed in ref. Citation8). Although retroviruses display considerable diversity, gRNAs have in common three open reading frames (ORFs). Overlapping gag and pol reading frames encode major structural (Gag) and structural-catalytic polyproteins (Gag-Pol). Gag-Pol is most typically expressed via programmed frameshifting which insures that it is produced at lower levels than Gag. The third ORF encodes the envelope protein (Env) expressed from a spliced RNA.Citation11
Despite commonality of structure and function, retroviral Gag proteins are not highly conserved in sequence. Gag domains processed into mature proteins common to most retroviruses include matrix (MA), capsid (CA) and a RNA binding domain (reviewed in ref. Citation12–Citation14). Gag-Pol is processed during maturation into Gag-derived species as well as protease (PR), reverse transcriptase (RT) and integrase (IN). For foamy viruses, the RNA-binding domain has an arginine-rich RNA-binding motif;Citation15 for alpha-, beta- and gamma-oncoretroviruses and lentiretroviruses the RNA-binding domain is NC. It contains one or two CX2CX4HX2C zinc finger (ZF) motifs embedded in a region enriched for charged basic and Asn and Gln residues (reviewed in ref. Citation8–Citation10). These domains are discussed in more detail below.
Retrovirus VLP assembly can be intracellular, as in the cases of betaretroviruses and foamy virus, or can occur on the plasma membrane, as in the cases of alpha- and gammaretroviruses and lentiviruses. VLPs or multimeric precursors, respectively, are translocated to the plasma membrane where they bud in areas enriched in lipid rafts associated with Env protein (reviewed in ref. Citation16 and Citation17). Within the context of the enveloped core, PR is activated and cleaves Gag and Gag-Pol polyprotein precursors into mature MA, CA, NC, PR, RT and IN and other virus-specific proteins, a process which is associated with rearrangement of the spherical immature core into its characteristic mature form. With the exception of foamy viruses,Citation18 cDNA synthesis prior to infection of the new host cells is limited to a single-stranded R-U5 complementary (c)DNA known as minus-strand strong-stop (-SSS). Within the new host cell, cDNA synthesis continues within a reverse transcription complex that is translocated along the microtubule network toward the nucleus. Finally, a preintegration complex is translocated into the nucleus and integration of the cDNA completes the cycle (reviewed in ref. Citation19).
LTR retrotransposon genome organization.
LTR retrotransposons have a similar genomic structure to that described above for retroviruses and encode major structural and catalytic proteins ().Citation1–Citation3 As used in this review, the term retroelement will refer to retroviruses and retrotransposons. Retrotransposons are typically expressed, similarly to retroviruses, from overlapping ORFs. Because the exemplary retrotransposons in this review will be mainly budding yeast S. cerevisiae Ty elements (reviewed in ref. Citation20 and Citation21), we use the terms GAG and POL which follow the budding yeast convention of italicized and capitalized “gene” names to refer to the structural and catalytic protein ORFs, respectively. Ty1 GAG and POL are also sometimes referred to as TYA and TYB, respectively. Some LTR retrotransposons, including gypsy in the Metaviridae family, encode Env proteins that mediate an optional extra-cellular replication cycle.Citation22 Thus, the classification of a retroelement as a retrotransposon may be more related to its ability to replicate intra-cellularly and its relatedness to other retrotransposons than to its inability to infect new host cells. One clear reflection of the intracellular lifecycle of retrotransposons compared to retroviruses is the simpler domain structure of Gag, which can be comprised simply of CA and RNA-binding domains.
Five major classes of LTR retrotransposons are cataloged by the International Committee on Viral Taxonomy (ictvonline.org). Of these elements, the Pseudoviridae (previously copia-Ty1) and Metaviridae (previously gypsy-Ty3) families are most similar to retroviruses according to alignments within the Pol domain.Citation1 POL ORF organization and sequence of Metaviruses (PR-RT-IN) are more similar to those of retroviruses than are those of Pseudoviruses (PR-IN-RT). There is surprising variability within families with respect to the type of RNA-binding domain. The Metaviridae family of LTR retrotransposons includes Errantiviruses (gypsy and ZAM); Metaviruses (Ty3, Tf1, SURL and Sushi); and Semtoviruses (Tas and Bel). The Pseudoviridae family includes Pseudoviruses (Ty1, 2 and 4); Hemiviruses (Ty5 and copia); and Sireviruses (SIRE1). However, the RNA-binding domains have diverged in complex ways. For example, Metavirus Tf1 and the Pseudovirus Ty1 lack the CX2CX4HX2C ZF, while the Pseudoviruses copia and Ty5 and the Metavirus Ty3 contain versions of this domain. Where it occurs, the motif is not as conserved among retrotransposons as it is among retroviruses (reviewed in ref. Citation23).
The TY3 Model
Ty3 genome and proteins.
The Ty3 Metavirus adheres to a subset of the general properties of retroviruses described above. It is 5.4 kb in length, comprised of 340-bp LTRs and overlapping GAG3 and POL3 ORFs (). It is transcribed into a R-U5-GAG3-POL3-U3-R polyadenylated RNA in which POL3 slightly overlaps the downstream U3 region. GAG3 and POL3 are translated into Gag3 and Gag3-Pol3 polyproteins that assemble into VLPs (reviewed in ref. Citation21). A high Gag3:Gag3-Pol3 ratio is maintained by programmed frameshifting.Citation24,Citation25 Similar to some retroviruses, including HIV-1 (),Citation26 and Rous sarcoma virus (RSV),Citation27 CA and NC are separated by a short and essential spacer (SP). The 290-aa Gag3 is processed into mature 206-aa CA, 26-aa SP and 57-aa NC.Citation28,Citation29 Gag3-Pol3 is processed into those proteins and PR, junction (J),Citation30 RT and IN.Citation29,Citation31 Because NC is encoded at the downstream end of GAG3, the coding region actually spans the frameshifting site so that a 76-aa NC protein is generated by processing of Gag3-Pol3 at SP-NC and at the amino-terminus of PR.Citation32 Ty3 NC is similar in size and charge to retroviral NC species (). Thus, Ty3 provides a relevant model in S. cerevisiae for studying aspects of NC function that might be conserved or divergent depending upon requirements for viral and retrotransposon lifecycles.
Experimental control of Ty3 expression and localization of RNA and protein products.
Because retrotransposons are endogenous they can accumulate in multiple copies in host genomes and confound studies with single experimentally-manipulated elements. In the case of Ty3 there relatively few native elements in common laboratory strains. In addition, strains were developed from which endogenous elements were deleted so that experimental Ty3 elements provide the sole source of Ty3 protein and gRNA.Citation33 Expression is naturally induced by exposure of cells to pheromone during mating.Citation33,Citation34 For some experimental applications, Ty3 elements are used in which the GAL1-10 upstream activating sequence is substituted for the upstream U3 region so that transcription can be artificially upregulated upon exposure to galactose as a carbon source.Citation35
Ty3 proteins are monitored in living cells by fusing the green or red fluorescent protein (G/RFP)-coding regions in-frame to the downstream end of GAG3 or POL3 or in fixed cells using antibodies against Ty3 CA, NC and IN and fluorescent secondary antibodies (reviewed in ref. Citation31 and Citation36). Intracellular localization of Ty3 RNA has been monitored by tagging individual RNAs with the RNA-binding site for the MS2 phage capsid protein and co-expressing MS2 fused to RFP or GFPCitation36 or by fluorescence in situ hybridization (FISH) (unpublished data). Examination of cells expressing Ty3 using microscopic imaging of fluorescently-tagged Ty3 protein or RNA shows that the majority of cells form clusters of Ty3 RNA and proteins. Immuno- and transmission electron microscopy (TEM) showed that these foci also contain clusters of VLPs. These clusters are associated with RNA processing (P)-bodyCitation36 and stress-granule proteins (Sandmeyer SB and Bilanchone V, unpublished data) and have been proposed to represent sites of intracellular assembly.Citation36 We refer to retrotransposon assembly foci as “retrosomes”.
Retroviral and TY3 NC Proteins
Retroviral NC function.
The structure and functions of retroviral NC have been the subject of several recent excellent reviews.Citation6–Citation10 The retroviral Gag NC domain or the mature NC engage in multiple aspects of the replication cycle. Activities include gRNA dimerization and packaging, Gag nuclear translocation, Gag multimerization, primer tRNA annealing, cDNA strand transfers, nonspecific reverse transcription suppression and integration enhancement. In addition to direct involvement of NC in these steps, NC mediates viral interactions with a remarkable assortment of host factors. The gamma retrovirus Moloney murine leukemia virus (MoMLV) NC has one ZF, but most other retrovirus NC domains have two copies (). The first of these is typically closer to the overall consensus: C1φ2XC4G5XXG8H9XXXD/E14C15. MoMLV also has a slight variation of the consensus: C1A2Y3C4K5XXG8H9XXXD14C15.
Ty3 NC proteins.
Mature Ty3 NC is processed from Gag3 (CA-SP-NC) and Gag3-Pol3. The mature 57 aa NC protein produced from Gag3 is similar to most retroviral NC proteins in size (MW = 6924.82) and charge (pI = 11.15). Processing of Ty3 Gag3-Pol3 produces an additional minor 76-aa species. However, there is no evidence that NC derived from Gag3-Pol3 has a unique function. Experiments in which the two Ty3 NC forms were provided from elements that were independently mutated showed that either source of NC was sufficient to support transposition, although relative stoichiometry of the assembled species was unrestricted in the experimental design.Citation32 The 57-aa NC is basic due to the presence of seventeen charged basic residues and nine Asn residues. It contains a single ZF motif. The Ty3 version is C1F2Y3C4K5XXG8H9XXXE14C15, which is similar to the retrovirus ZF consensus, but has aromatic residues at both the second and third positions and lacks the highly-conserved G5. Interestingly, The aromatic residue at position 3 and the absence of G5 in the MoMLV NC ZF are similar to the Ty3 variation. This review will focus on the functions of NC related to stages of the retrotransposon replication cycle having to do with RNA.
Initiation of Assembly
Gag assembly domains.
Three major domains have been implicated in retroviral particle assembly: first, the MA M domain, including an amino-terminal acylation site and concentration of basic residues, both of which promote membrane association; second, a domain at the carboxyl-terminal end of CA, in the case of HIV-1 including SP1, which participates in Gag-Gag interactions; and third, the NC domain which mediates interaction with the RNA scaffold and thereby indirectly promotes multimerization.Citation37–Citation40 Of these, in the case of HIV-1, the CA-SP1 domain contributes to assembly primarily through inter-Gag interactions.Citation41 Retrovirus Gag also contains one or more late (L) motifs of several amino acids that provide for interactions with components of the cellular endosomal sorting complexes required for transport (ESCRT). These interactions are required for envelopment of the assembled particle during budding (reviewed in ref. Citation16).
Ty3 CA modeling suggests that, similar to retroviral CA, it is composed of an amino-terminal domain (NTD) bundle of alpha helices (residues 2–139) and smaller carboxyl-terminal domain (CTD) bundle (residues 148–207).Citation42 Two-hybrid assays showed that the NTD interacts both with itself and with the CTDCitation43 and particle formation is sensitive to mutations in the first 100 residues of the NTD.Citation42 Mutations in some but not all residues in the conserved ZF profoundly disrupt VLP formation,Citation44 consistent with the model that specific Gag3 binding to RNA is important for the correct particle structure. Ty3 particles visualized by AFM are roughly spherical and have broadly-distributed diameters of approximately 42 nM. The distribution of surface capsomeres show elements of five-fold and six-fold symmetry consistent with an icosahedral-like structure.Citation45
Cytoplasmic trafficking and collection of retroviral VLP components.
The natural overlap of retroelement transcription, translation and assembly complicates clear definition temporally and spatially of the point at which assembly initiates. Nonetheless, evidence supports the general view that components concentrate at subcellular locations as part of the assembly process.Citation46 Retrovirus assembly progresses through translation of the RNA to produce Gag and Gag-Pol; commitment of RNA to assembly; concentration of gRNA, Gag and Gag-Pol at the pre-assembly or assembly site; and assembly and budding (reviewed in ref. Citation6, Citation16, Citation46 and Citation47).
Multiple mechanisms cooperate to result in assembly of retroviral gRNA into particles.Citation48 Because retroviral RNAs templating Gag and Gag-Pol synthesis or those destined to become gRNAs are unspliced, they require specialized nuclear export pathways.Citation49 This role can be fulfilled by either retroviral proteins, such as HIV-1 Rev, that bind to specific sequences in the gRNA and act as adapters for cellular export proteins (reviewed in ref. Citation50 and Citation51), or by a viral constitutive transport element (CTE), such as that of MPMV, in the gRNA that is recognized directly by cellular export factors.Citation52 There could also be designation of gRNA in the nucleus. The gammaretrovirus MoMLV gRNA is derived from a pool of newly-made transcripts separate from translating RNAs.Citation53 Consistent with that, these gRNAs are more likely to be co-packaged with RNAs transcribed from nearby loci, suggesting that dimerization could be initiated in the nucleus.Citation54 In the case of RSV, Gag shuttles through the nucleus and interruption of this cycle decreases gRNA packaging. This Gag nuclear localization is mediated by MA domain interactions with importin-11 and NC domain interactions with importin-alpha/beta.Citation55,Citation56 For other retroviruses, including HIV-1, the translating pool of viral RNAs contributes to gRNA.Citation48 However, for HIV-1, redirecting nuclear export from one pathway to another can disrupt packaging.Citation57 These findings suggest that even retroviral RNAs that are within the translating pool could be designated as available for packaging soon after transcription. Because there is not a requirement for packaging in cis, untranslated gRNA could be packaged in principle and Gag must be able to bind gRNA in trans with specificity. In vitro experiments demonstrated that HIV-1 Gag can negatively autoregulate translation, offering an elegant mechanism through which retrovirus RNAs could transition from translation template to gRNA.Citation58 These examples do not exclude the possibility of a temporal transition in gRNA designation. For example, if in the course of an infection accumulation of unbound Gag resulted in Gag nuclear entry and gRNA capture.
As alluded to above, psi sequences in the gRNA mediate selective Gag binding. When ZFs of retroviral NC proteins were swapped, the source of the ZF determined gRNA binding specificity and mutations in the ZF disrupted gRNA packaging.Citation59,Citation60 For most retroviruses for which psi is defined, it consists of several hundred-nt containing a few stem-loops near or within the gRNA 5′ UTR (reviewed in ref. Citation61). Psi sequences for which function is understood contain a dimerization initiation site (DIS), which can nucleate dimer formation via a “kissing loop” interaction. NC promotes an extension of this interaction by chaperoning re-pairing of the strands in the monomeric gRNA stems to form mature intermolecular duplexes in the dimeric gRNA. NMR studies have characterized the MoMLV and HIV-1 NC ZF complexed with psi stem loop regions (reviewed in ref. Citation62 and Citation63). In the case of MoMLV, high-affinity NC binding sites become accessible in the extended structure, suggesting the basis for stabilization of the dimer. Evidence that NC loss-of-function mutations reduced particle formation and that such mutations were rescued by substitution of a leucine zipper domain for the ZF support a model in which the ZF in mediating Gag binding to a RNA scaffold, stabilizes weaker inter-Gag interactions.Citation39,Citation64
Examination of cells over the course of a retrovirus infection shows that assembly is initiated prior to delivery of Gag to the plasma membrane. Fluorescence resonance energy transfer (FRET) experiments visualize intermolecular Gag interactions within small cytoplasmic foci.Citation65,Citation66 In addition, pulse labeling and examination of multimeric species over a time course shows formation of Gag multimers prior to membrane delivery.Citation67–Citation69 Intracellular Gag trafficking appears to be dependent upon gRNA, as deletion of psi in the case of MoMLVCitation70 and HIV-1,Citation71 resulted in loss of trafficking to the cell surface and to the perinuclear microtubule organizing center (MTOC), respectively. As would be predicted if interaction depended upon the ZF for specificity, mutations in HIV-1 ZF also resulted in a diffuse pattern of Gag.Citation72 Together these experiments indicate that specific binding by the NC domain of Gag is required for formation of cytoplasmic assembly intermediates.
There is evidence that HIV-1 protein and RNA assembly intermediates collect at the perinuclear MTOC. Fluorescence and pulse chase experiments demonstrate collection of Gag multimers and gRNA at this site.Citation67,Citation71,Citation73 Treatment of cells with latrunculin disrupts this localization, implicating the actin cytoskeleton in formation of pre-assembly foci.Citation71 Other data indicate that actin associates with HIV-1 Gag via the NC domain.Citation74–Citation77 Localization likely promotes further multimerization. In addition, chaperonins are associated with the MTOC. In the case of betaretroviruses, which assemble into VLPs in a peri-MTOC compartment prior to endocytic trafficking to the cell surface, Gag has been shown to interact with chaperonins normally associated with tubulin assembly.Citation78 Although HIV-1 does not fully assemble at the MTOC, chaperonins might modulate multimerization. The assembly factor ABCE1 associates at early stages of multimerization with Gag and this interaction is also mediated by NC.Citation69,Citation79 HIV-1 RNA contains binding sites for the ribonucleoprotein (RNP) component, hnRNP A2. Knockdown of hnRNP A2 in HIV-1 producing cells causes accumulation of HIV-1 RNA in the nucleus and in the vicinity of the MTOC.Citation73 Thus, accumulation of assembling complexes might also result in recruitment of newly-transcribed RNA for assembly. Finally, proximity to the MTOC offers access to the microtubule network and endocytic trafficking to the cell surface where assembly and/or budding occur (reviewed in ref. Citation49).
Collection of Ty VLP proteins in retrosomes.
Retrotransposons assemble through a contracted version of the retrovirus pathway: translation of the RNA to produce Gag and Gag-Pol; commitment of RNA to assembly; and concentration and assembly of gRNA, Gag and Gag-Pol. Similar to retroviruses, the Ty3 retrotransposon translates template RNA into Gag and Gag-Pol which are observed in multiple foci shortly after induction.Citation31,Citation36 However, in contrast to retroviruses, these Ty3 foci coalesce within a few hours into one or two large foci or retrosomes per cell. VLPs are primarily but not exclusively observed in the foci, and Gag assembly mutants show defects in focus formation.Citation42 Therefore foci are presumed to represent Ty3 assembly sites. Ty1 retrotransposons form similar concentrations of Ty RNA, protein and VLPs.Citation80–Citation82 Ty1 and Ty3,Citation36 elements appear to share certain “host” components of these clusters, but not others, indicating that Ty1 and Ty3 clusters are likely distinct. However, because representatives of both major classes of LTR retrotransposons in S. cerevisiae assemble particles within foci of concentrated components, we refer to both classes of retrotransposons assembly clusters as retrosomes. In yeast the spindle pole body acts as the microtubule organizing center. Despite a perinuclear position in many cells, Ty3 retrosomes do not consistently localize in the vicinity of the spindle pole body (Sandmeyer SB and Beliakova-Bethell N, unpublished data). Thus, the retrosome is probably functionally distinct from the peri-MTOC concentration of assembling betaretroviruses or lentivirus intermediates.
Collection of Ty3 RNA in retrosomes.
Concentration of Ty3 RNA in retrosomes requires both cis-acting and trans-acting functions. Although psi has not been mapped for Ty3, analysis of cells expressing helper Ty3 elements and a HIS3-tagged donor Ty3 showed that the donor 5′ Ty3 UTR and downstream PPT-U3-R sequences were sufficient to allow retrotransposition of the HIS3 marker. Thus, these Ty3 sequences are sufficient for packaging, reverse transcription and integration.Citation24 Expression of Ty3 RNA results in rapid concentration of Ty3 protein and RNA into retrosomesCitation36 and protection of the RNA from nuclease digestion, indicating that it is packaged. In contrast, similarly expressed control RNAs do not form such clusters and are not protected from digestion, even in cells expressing Gag3. It was anticipated that sequences required for retrotransposition would include those required for localization to retrosomes and for Gag3-mediated assembly. When chimeric RNAs were tested, either the Ty3 5′ UTR and 3′ PPT-U3-R or GAG3-POL3, flanked by non-Ty3 UTRs (but not GAG3, flanked by non-Ty3 UTRs), was sufficient for RNA localization to retrosomes in cells expressing Gag3 (SBS, Beliakova-Bethell N, KAC, Bilanchone V, manuscript in preparation). However, sequence flanked by the Ty3 5′UTR and 3′PPT-U3-R was packaged, whereas the GAG3-POL3 RNA flanked by non-Ty3 UTRs was not. Thus, Ty3 RNA promotes assembly at two stages. In the first stage, association is specific, but is mediated by the 5′ UTR and 3′ PPT-U3-R or POL3 and allows for cytoplasmic coalescence of Gag3 and RNA with retrosome components. In the second stage, sequences within the Ty3 5′ UTR and PPT-U3-R are recognized by Gag3 and gRNA is assembled into VLPs. Retrosome localization does not insure RNA assembly into Ty3 VLPs.
Ty3 retrosomes contain P-body and stress granule components.
The dramatic formation of retrosomes observed in cells expressing Ty3 prompted investigation of the cytoplasmic components with which they are associated. Because P-body proteins had been implicated in retrotransposition by genomewide screens of mutants defective in Ty1,Citation83 and Ty3,Citation84 the association of a subset of these with Ty3 components was explored. In S. cerevisiae, proteins identified in P-body complexes include proteins implicated in translation repression (Dhh1, Pat1 and Ded1), decapping (Lsm1, Edc1, Dcp2), RNA unwinding (Dhh1 and Ded1), 5′ to 3′ RNA degradation (Xrn1/Kem1), and non-sense-mediated decay (NMD) (Upf1-3).Citation85,Citation86 Dhh1, Pat1, Edc3, Dcp2 and Xrn1/Kem1 GFP reporter fusions co-localize with fluorescently-tagged Ty3 protein and Lsm1, Dhh1 and Xrn1 are essential for wt levels of retrotransposition.Citation36,Citation84 Sequestration of assembling particles with RNA degradation factors is counterintuitive. However, in yeast P bodies are necessary for formation of stress granules in which RNA can be sequestered during stress and then returned to polysomes,Citation85,Citation87,Citation88 and recent work shows that decapping-dependent RNA degradation occurs co-translationally.Citation89 In addition, mass spectrometry analysis of proteins co-purifying with Gag3 complexes identified a subset of stress-granule proteins that have been localized to retrosomes by fluorescence microscopy (Bilanchone V, unpublished results). Thus, P-body and stress-granule proteins could cooperate with Gag3 to create an assembly environment sequestered from translation and degradation. The association of retrotransposon assembly with RNA granules would make considerable sense. P-body translation repression functions would promote transition from translation to assembly while stress-granule functions would include proteins involved in stabilizing or folding other proteins, binding RNA and initiating translation.Citation90,Citation91 Such factors could provide chaperones for assembling Ty3 proteins as well as concentrating initiation factors to recruit primer initiator tRNAMet (tRNAiMet), to the retrosome.
Several results indicate that there is likely to be physical association between Ty3 proteins and RNA and P-body and stress-granule proteins. Mutations in Ty3 CA that disrupt Ty3 particle formation have effects on P-body protein patterns. Two CA mutations that profoundly disrupted Ty3 assembly dissociated Ty3 Gag-Pol-RFP and Dhh1-GFP fluorescence. A mutation in CA that caused formation of fibrillar Gag3-Pol3-RFP structures throughout cells caused a similar reorganization of Dhh1-GFP and Xrn1/Kem1-GFP reporters.Citation42 The DEAD-box helicase Dhh1 tagged with GST co-precipitates with Gag3, although it is not known if this represents a direct or indirect interaction (Beliakova-Bethell N, Clemens K, unpublished data).
The role of retrosomes in retrotransposition is not likely to be simple; in spite of clear evidence that P-body and stress-granule components contribute in positive ways to retrotransposition, other data point to a role for retrosomes in limiting transposition. For example, a Gag3 amino-terminal Ala substitution mutation decreased cluster size, but increased transposition dramatically (Sandmeyer SB, Zhang M, Bilanchone V, unpublished data).
Model for Ty3 transition from template to genomic RNA.
An intriguing problem in the case of translated retroviral genomes is how the transition of the RNA from translation template to gRNA is controlled. A mutation in the Ty3 translation initiation AUG of Ty3 elements marked with a HIS3 insert essentially eliminates retrotransposition in cells expressing helper Ty3 (Sandemeyer SB, Beliakova-Bethell N, unpublished data). Although the mutant Ty3 RNA level is low, if gRNA were delivered directly from the nucleus without translation, this mutation would not alter assembly into particles. The reduction in transposition suggests that at least some amount of Ty3 gRNA is derived from the translated pool. The 5′ and 3′ ends of translating RNAs are bound by translation initiation factors and poly(A) binding proteins, respectively and are bridged by eIF4G (reviewed in ref. Citation92). Circularization is proposed to protect the ends of the translating RNA and promote delivery of the reinitiating 60S subunit to the initiation site. However, retroelement RNA, which represents both the template and the genome, is subject to special constraints. Because programmed frameshifting releases ribosomes from retroelement template RNAs upstream of the 3′ ORF, retroelement gRNAs have relatively low downstream ribosomal occupancy. This could attenuate translation reinitiation on retroelement RNA and make it susceptible to P-body protein mediated translational repression and aggregation. Three observations make this particularly relevant to yeast LTR retrotransposons. First, tRNAiMet is the reverse transcription primer used by Ty1, Ty3 and Ty5 yeast LTR retrotransposons and is observed in the vicinity of Ty3 retrosomes (Bilanchone V, unpublished results). Second, Ty3Citation36 retrosomes include P-body translational repressors (e.g., Dhh1 and Lsm1).Citation85,Citation87,Citation88,Citation93,Citation94 Third, Ty3 and Ty1 require subsets of P body-proteins for retrotransposition.Citation81,Citation82 We speculate that the Gag3 binds to psi sequences in the retroelement gRNA and interferes with translation initiation, increasing their susceptibility to binding by P-body translation repressors and thereby nucleating retrosome coalescence.
Effects of substitution mutations in conserved residues of the Ty3 ZF.
On the basis of effects of mutations in the MoMLV,Citation95 RSVCitation96 and HIV-1,Citation97 ZF, mutations in Ty3 ZF might have been expected to leave particle assembly intact and result in nonspecific RNA packaging. However, substitution of three conserved Cys residues and one His residue in the single ZF of Ty3 NC showed an unexpected range of phenotypesCitation44 and dramatically decreased assembly. A model in which the ZF acts as a modular structure, predicts that substitution of any Cys or the single His residue within the Ty3 ZF would similarly disrupt zinc chelation and therefore function. Surprisingly, substitution of Ala for conserved ZF residues in Ty3 NC (C1, C4, H9 and C15) produced graduated effects, progressing from most severe disruption of assembly at the amino-terminal end (C1A) to least severe at the carboxylterminal end (C15A). C1A mutant Gag3 localized to the nucleus where it formed electron-dense aggregates and loose spheres. In contrast, H9A and C15A mutants formed electron-dense “balls” of Gag3 with significant numbers of extra-nuclear particles resembling VLPs. Similarly, Ty3 RNA foci were more severely diminished in cells expressing the C1A mutant than in cells expressing the C15A mutant. The effects of these mutations argued that a region overlapping the ZF plays an important role, not only in assembly, but in cytoplasmic retention of Gag3. Mutant Gag3-Pol3 tagged with GFP did not localize to the nucleus and in the less severe H9A and C15A mutants, some particles were observed both inside and outside of the nucleus. In cells expressing wt Ty3, nuclear Gag3 is not detectable. However, an interesting possibility raised by these results is that in the absence of RNA binding, wt Gag3 might traffic into the nucleus to recruit newly-made RNA for packaging.
Role of the basic NTD of Ty3 NC.
Retrovirus NC function as a RNA chaperone depends upon a positive-charge bias.Citation8–Citation10 Ty3 NC is similarly positively charged with 17 basic residues. A series of Ala-scanning mutants in which two or three charged residues within a window of five aa were changed to Ala was examined to determine the contributions of basic residues to VLP morphogenesis.Citation44 None of the double mutations affected transposition and only two of the four triple mutations resulted in partial transposition defects. The stronger of these changed charged residues within the ZF to Ala and resulted in a 50% decrease in cDNA production. Overall, the effects of substituting local sets of charged residues in the Ty3 NC protein indicates that they do not individually perform essential functions in retrotransposition.
In order to test for roles of the basic NTD and NC itself, a mutant having a deletion of the basic NTD (ΔNTD, residues 236 to 265 including 10 basic residues), and a mutant having a NC domain deletion (ΔNC aa 237 to 281) were tested for effects on retrotransposition.Citation44 ΔNTD and even ΔNC mutations permitted CA-SP processing, formation of Gag3 matrices, NC function is not required for some level of particle assembly and maturation. However, ΔNTD or ΔNC severely disrupted transposition and abrogated cDNA synthesis. These mutants were evaluated for localization with P-body proteins, VLP assembly and RNA packaging. Overall, deletion of the entire NC domain had similar, but more dramatic effects than deletion of the basic NTD. These reduced, but did not eliminate Gag3-Pol3-RFP foci, presumably based on CA domain interactions. These foci had reduced association with P-body components. Similar to the ZF mutants, ΔNTD and ΔNC Gag3 partitioned into the nucleus. TEM analysis showed that within the nucleus, ΔNC Gag formed a striking, tightly-packed matrix array of particles without electron-dense centers. However, ΔNTD formed electron-dense, fuzzy balls that were larger than normal VLPs. Both classes of mutations eliminated gRNA protection, consistent with a central role for the basic domain of Ty3 NC in RNA association.
Gag3 nuclear localization activity in Ty3 NC.
The results of experiments described above indicated that Gag3 remains predominantly cytoplasmic due to an activity that maps to a region overlapping the ZF. In addition, they suggest either that Gag3 not engaged with RNA can diffuse or that it can be actively translocated into the nucleus. In order to map domains of Gag3 with NLS activity, Gag3 and CA, CA-SP, CA CTD, CA NTD, SP-NC and NC were expressed fused to tandem repeats of the GFP-coding sequence (2xGFP).Citation98 NC-2xGFP concentrated in the nucleus and SP-NC-2xGFP showed diffuse fluorescence in the cytoplasm and nucleus. Other fusions formed foci some of which appear nuclear or perinuclear.Citation98 As described above even ΔNTD and ΔNC mutant Gag was found in large nuclear aggregates, although localization of those deletion mutants could have occurred by diffusion and retention of aggregates.Citation44 RSV Gag is actively transported into the nucleus and exported.Citation55,Citation99,Citation100 Interruption of this pathway does not block protein expression, but causes decreased gRNA packaging,Citation100 supporting the model that RSV Gag captures gRNA in the nucleus. Although wt Ty3 Gag3 does not accumulate in the nucleus to detectable levels, evidence that Gag3 contains NLS activity and that mutations that interfere with Gag3 binding to Ty3 RNA result in Gag3 nuclear accumulation would support a two-phase model. In the first phase, Gag3 associates with Ty3 gRNA in the cytoplasm, but in the second phase, unbound Gag3 accumulates and then translocates into the nucleus where it associates with newly-transcribed Ty3 RNA. This model would require nuclear export of Ty3 Gag which has not yet been demonstrated.
Interactions of NC with Related Host Factors during Assembly
Given the size and structural constraints of the retroviral NC domain, it participates in an amazing spectrum of assembly and replication functions. Interacting proteins include ones with functions upon which the virus is dependent, as well as at least one host restriction factor. Examples of interactions occur throughout retrovirus assembly. As described above, the NC domain mediates RSV Gag interactions with importins alplha/beta.Citation55 Evidence suggests that the NC domain could also play a role in formation of assembly intermediate foci for HIV-1. It has been known for some time that actin interacts with HIV-1 GagCitation76,Citation101,Citation102 and recent experiments showed that the actin cytoskeleton is required for localization of Gag and gRNA to the MTOC preassembly site.Citation71 The NC domain also mediates interactions with assembly factors. The contribution of staufen required for gRNA packaging depends upon the NC domain.Citation103,Citation104 The HIV-1 assembly factor ABCE1 which promotes early multimerization of Gag interacts with Gag via residues close to the ZF motif.Citation79 Nucleolin, a ribosomal assembly factor interacts with the CA-NC domain junction and mutations at this junction disrupt MLV assembly.Citation105 ESCRT complex budding factors associate with Gag and this interaction depends upon interactions within Gag late domains—so called because disruption results in a late cycle budding defect. The host factor ALIX/AIP1 mediates a subset of those interactions.Citation106 Results from several groups have recently demonstrated that host late functions are redundant with some activities of NC. For example, overexpression of late-acting ESCRT components known to interact with ALIX/AIP1,Citation107 or substitution of NC with the leucine zipper domainCitation108 suppress a subset of effects of certain NC mutations on budding. Finally, HIV-1 host restriction factor ABPOBEC3G, assembles into particles via RNA-dependent NC interaction.Citation109–Citation111 Thus, NC is a multifunctional assembly domain that exploits interactions with host factors and is in turn targeted by host restriction.
An intact NC domain in Gag3 is required for development of Ty3 retrosomes which are potentially associated with P-body proteins. Although certain P-body functions clearly have positive effects on retrotransposition, certain Ty3 mutant phenotypes suggest that large retrosomes are repressive to Ty3 transposition. Thus for Ty3, P-body functions are associated with positive and negative effects. At least some P-body functions are also implicated in HIV-1 assembly. For example, staufen, is associated with P bodies.Citation112 HIV-1 infectivity is sensitive to non-physiological levels of Mov1, a RNA helicase associated with P bodies.Citation113 Finally, the restriction factor APOBEC3GCitation81,Citation114 and miRNA antagonists of HIV-1,Citation115,Citation116 localize with P-body components. Thus, both Ty3 and HIV-1 interact with subsets of P-body proteins. Because of its chaperone activity, NC is likely to play an important role in mediating access of these factors to the gRNA.
Processing and Activities of Mature NC in Reverse Transcription
SP-NC context and maturation.
In spite of the conservation of NC ZF sequence and overall basic composition, the context of NC varies among retroviruses. In some retroviruses, including RSV, HIV-1 and equine immunodeficiency virus, CA and NC are separated by a short SP domain. In other retroviruses, including MoMLV, CA and NC are adjacent domains. Modeling based on electron cryomicroscopy of immature HIV-1 particles predicts formation of a bundle of SP alpha helices underlying the CA hexamers. This configuration is proposed to promote capsomere formation.Citation41 In the case of HIV-1, cleavages occur in the order NC-P6, MA-CA, SP-NC and CA-SP.Citation12,Citation117 Thus, release of NC precedes separation of CA from the underlying SP bundles. Disruption of the SP-NC cleavage blocks condensation of NC and gRNA into an electron-dense particle core.Citation26
The Ty3 NC domain, similar to the HIV-1 NC domain, in the polyprotein precursor is flanked by a short SP domain. However, in contrast to HIV-1, Ty3 SP is highly acidic. Mutants in which Ty3 SP-NC processing is blocked, similar to HIV-1 mutants for which processing is blocked, fail to form electron-dense cores (Sandemeyer SB, Clemens KA, in press). In contrast, Ty3 SP deletion mutants form particles and make cDNA, but fail to retrotranspose. The strong negative charge in SP might allow it to interact with the basic domain of NC during assembly. A mutant in which acidic residues were substituted with Ala failed to form retrosomes and assemble. TEM analysis showed a limited number of cells with partially-formed, VLP-like structures that lacked the tight packing observed in wt or PR- mutant retrosomes. These results suggest that Ty3 SP is not absolutely required for assembly, but if present, must be negatively charged. In the case of human T-cell lymphotrophic virus-type 1 (HTLV-1) acidic residues located in the carboxyl-terminal domain of NC are associated with lower rates of NC dissociation from nucleic acid substrates and therefore less effective chaperone activity than some other retroviral NC proteins.Citation118,Citation119 It was proposed that intermolecular actions of the acidic and basic domains of HTLV-1 NC promote packing on the RNA. In the case of Ty3 Gag3, SP acidic residues appear to be required for proper assembly, but intermolecular interactions between SP and NC domains might similarly promote stable assembly of Gag3 onto the gRNA.
Ty primer annealing and gRNA circularization.
Retroviral Gag exhibits higher nucleic acid binding and aggregating activity than processed NC, consistent with its role in packaging RNA into the VLP.Citation120 However, subsequent to proteolytic maturation, NC performs important chaperone functions throughout reverse transcription. Because of the critical role of NC in assembly, much of what we know about its activities in reverse transcription is based upon in vitro experimentation. NC plays supporting or key roles in RNP complex formation, suppression of premature reverse transcription in producer cells, melting of internal RNA structures and annealing of primer tRNA and cDNA intermediates, (reviewed in ref. Citation8–Citation10). Less work has been performed with Ty3 NC than retroviral NC. However, in vitro assays indicate that it functions in at least a subset of these roles.
One of the most fascinating aspects of LTR retrotransposon replication is the diversity of reverse transcription priming strategies even within retrotransposon families. The Metavirus Ty3 as well as the Pseudoviruses copia, Ty1,Citation121,Citation122 and Ty5,Citation123 use tRNAiMet as the primer tRNA. However, Ty5 and copia prime from the 3′ end of a tRNAiMet internal cleavage product.Citation121,Citation122 The Metavirus, Tf1, self-primes from the 3′ end of an internal cleavage product of Tf1 gRNA.Citation124 Copia, Ty5 and Ty3 have ZF-containing NC proteins, but Ty1 and Tf1 do not. A basic peptide from the carboxyl-terminal domain of Ty1 Gag has RNA chaperone activity. This peptide and Ty3 NC can anneal primer tRNAiMet to the respective retrotransposon gRNAs in vitro.Citation125 Thus, the presence of ZF-containing NC is not correlated with the identity of the primer species. This convergence suggests that for retrotransposons the choice of the translation initiator tRNA as primer is driven by some specific functional benefit.
Although Ty3 NC and HIV-1 NC both have primer-annealing activity, there are interesting differences in the products of these reactions and the resulting reverse transcription templates. Early studies of Ty3 identified an 8-nt sequence 2 nt downstream of U5 that is complementary to tRNAiMet.Citation126 Despite the modest complementarity, effects of mutations in tRNAiMet argued convincingly that it was the primer.Citation122 Further inspection revealed additional complementarity to the primer within downstream U3.Citation127 The bipartite complementary sequence was required for tRNAiMet priming of -SSS cDNA synthesis by HIV-1 or Ty3 RT in vitro.Citation127,Citation128 Similarly, the Ty1 primer building site (PBS) initially identified as a 10 nt sequence complementary to the 3′ end of primer tRNAiMet was shown to have additional short segments of complementarity required for -SSS production. In contrast to Ty3, these were contained within the 5′ end of the gRNA.Citation129,Citation130 Remarkably, tRNAiMet has a 12 nt GC-rich palindromic sequence at the 5′ end which is unpaired when the tRNA is annealed to the Ty gRNAs. This strand of the tRNAiMet stem is necessary for NC-mediated formation of Ty3 dimers in vitro.Citation127
Ty3 NC promotes formation of Ty3 RT ribonucleoprotein complexes,Citation128 primer tRNAiMet annealing to Ty1Citation125,Citation127 and Ty3 template RNAs,Citation127 and tRNAiMet-primed reverse transcription of mini-Ty3 RNA to produce -SSS cDNA intermediates.Citation127 In addition to performing these functions on mini-templates, Ty3 NC anneals HIV-1 primer tRNALys3 to HIV-1 RNACitation127 and can promote dimerization of heterologous 5′ HIV-1 RNACitation127 and Ty1 RNAs in vitro.Citation125 Deletion mutation analysis indicates that the basic domain is more important for primer annealing, gRNA dimer and -SSS synthesis than is the ZF.Citation128
A key step assisted by NC in reverse transcription of retroelements is the transfer of the nascent -SSS cDNA template from the 5′ end of the gRNA to the 3′ end to allow extension of the minus-strand cDNA. Re-pairing of the R region of the -SSS chaperoned by NC has been proposed to mediate this transfer for retroviruses.Citation5 Most Ty3 transcripts would have a R sequence of about 50 nts. This fits within the range of retroviral R (e.g., RSV = 21 nts; MLV = 68 nts; HIV-1 = 98 nts). Nonetheless, because experiments showed that the Ty3 bipartite (5′ and 3′) PBS was required for -SSS, it seems likely that tRNAiMet bridges the 5′ and 3′ ends of the gRNA and so promotes strand transfersCitation127(). Despite the very strong appeal of this model, direct testing in vivo of the requirement for a bipartite primer linker for strand transfer is complicated by the epistatic requirement of even Ty3-SSS synthesis for bipartite primer annealing. This would not be the case for Ty1 where the multipartite PBS is in the 5′ end (). Efforts to develop an in vitro reverse transcription system for that element, however, identified a region downstream of the Ty1 PBS with 14 nt of complementarity to a sequence in the U3 that was required for efficient initiation of -SSS DNA.Citation131 Thus, at least two mechanisms to circularize the genome in addition to the R sequence overlap are represented in budding yeast retrotransposons.
Two interesting discoveries suggest that long-range interactions between the ends of gRNA in addition to those mediated by the R region may extend beyond the yeast retroelements. First, structural analysis of HIV-1 identified a phylogenetically-conserved complementarity within a 600–700 nt region within Gag and a region of 123 nt at the downstream end of the HIV-1 gRNA. Interaction of these sequences was also proposed to enhance strand transfersCitation132 (). Interestingly, a region of U3-R that contains extensive complementarity to HIV-1 primer tRNALys3 has also been shown to promote strand transfer.Citation133,Citation134 Finally, there is a possibility of covalent links between the 5′ and 3′ ends of retroelements that contribute to replication. Ty1,Citation135 Ty3 elementsCitation84 and later HIV-1,Citation136 were shown to require the lariat debranching enzyme (yDBR1) for replication. Evidence in both retrotransposon and retrovirus systems has suggested the existence of lariat forms in which gRNA would be circularized.Citation137,Citation138 However, conclusive demonstration of the role of such a branch point has been elusive.Citation139 Considering the ongoing function-structure analyses, as well as possibilities for high definition insights into in vivo RNA structuresCitation140 coupled with approaches to studying host gene interactions with NC, insights into the dynamics of the strand-transfer process are likely to be forthcoming..
Summary and Perspectives
The NC domain, both in the context of Gag and as a mature protein, contributes in critical ways to retroelement replication. Retrovirus NC has long been known to participate in gRNA recognition, dimer stabilization, packaging, primer annealing and cDNA strand transfers. New functions continue to be discovered including roles in multimerization, suppression of premature reverse transcription and mediation of localization and other complex interactions with the host cellular environment. Studies in the Ty3 Metavirus retrotransposon system have shown that retrotransposon NC is likely to perform versions of these functions tailored to the intracellular environment of the retrotransposon lifecycle. These include targeting gRNA and protein to retrosome assembly sites in association with P-body and stress-granule proteins, controlling cytoplasmic versus nuclear localization of Gag3 and annealing a multipartite tRNA primer that both circularizes and dimerizes the genome. Key aspects of retrovirus and retrotransposon NC functions remain to be resolved. Continued investigation is motivated by the potential of retrotransposons to impact the function of eukaryotic genomes and the potential of NC as an anti-retroviral therapeutic target. Questions posed by current models will continue to stimulate future research regarding this fascinating multi-functional protein. Is retrosome gRNA capture mediated by P-body translational repressors loading onto the translating RNA? Is a flanking acidic domain that modulates basic NC domain activity a common feature of retroelement replication? Is nuclear entry and gRNA capture a default pathway for unbound Gag? Is the genome circularization observed for Ty and Tf1 elements and HIV-1 a widespread feature of retroelements? Finally, and perhaps most interestingly, what are the triggers for transitions among these functions and what controls the dynamic interactions of NC with host proteins?
Figures and Tables
Figure 1 Ty3 and HIV-1 genome organization. (A) Ty3 genome and Ty3 RNA features. GAG3 encodes Gag3 containing capsid (CA), spacer (SP) and nucleocapsid (NC) domains; POL3 is translated as part of Gag3-Pol3 via a programmed frameshift and encodes protease (PR), junction (J), reverse transcriptase (RT) and integrase (IN) domains. Domains are processed into mature proteins by Ty3 PR. Ty3 RNA is capped, polyadenylated, and contains a bipartite reverse transcription primer tRNAi-Met binding site (PBS) in the 5′ leader sequence and in the U3 region of the transcript. Boxes at the ends of the genes indicate regions represented uniquely in the 5′ (U5), and 3′ (U3) ends of the RNA. The repeated (R) sequence is present at both ends. (B) HIV-1 genome. Gag encodes matrix (MA), CA, SP1, NC, SP2 and p6; pol encodes PR, RT and IN domains; a spliced RNA contains env which encodes envelope (Env); and vif, vpr, vpu, nef and tat are HIV-specific proteins encoded by spliced RNAs. Domains within retroviral Gag and Gag-Pol are processed by PR into mature species. U5, U3 and R are as in (A).
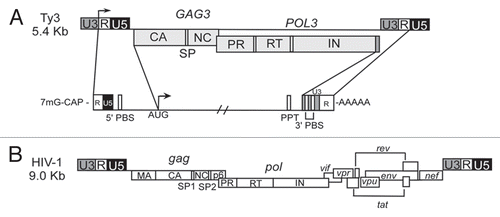
Figure 2 Ty3 and retroviral nucleocapsid proteins. Full-length sequences of NC proteins containing one or two conserved ZF motifs from the retrotransposon Ty3 and several retroviruses. Red, basic aa residues; green, conserved residues within ZF motifs depicted coordinating Zn2+. Sequence accession numbers: Ty3 AAA98434.2; MLV, J02255; HIV-1, AF324493; HIV-2, AAA76840.1; HTLV-1, AAB20767.1; RSV, AAC82560.1; and EIAV, AAA43011.1.
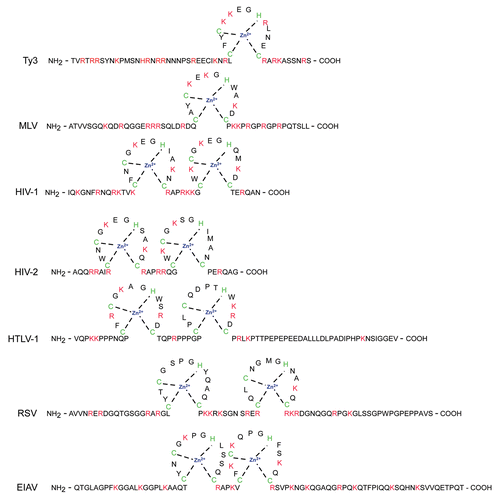
Figure 3 Circularized genomic RNA structures proposed for retroelement genomes with annealed primer tRNAs. (A) Ty3 RNA annealed to tRNAiMet. Ty3 RNA contains a bipartite primer binding site (PBS), comprised of regions in the 5′ leader and U3 of Ty3 RNA complementary to sequences in the 3′ acceptor end, TΨC arm and D arm of primer tRNAiMet.Citation127 Circularization is proposed to promote -SSS transfer. In vitro NC mediates dimerization of Ty3 gRNA dependent upon annealed tRNAiMet molecules paired via palindromic sequences at the 5′ ends of each tRNA. (B) Ty1 tRNAiMet dimerization. Ty1 RNA contains a multipartite tRNAiMet PBS in the upstream end that includes the originally-described PBS, Box 0, Box 1 and Box 2.1.Citation130 In vitro Ty1 dimerization is mediated by Ty1 RNA-binding protein via the palindromic sequences in the primer as described in (A).Citation125,Citation127 Circularization is dependent upon palindromic sequences in the 5′ (CYC5) and 3′ (CYC3) regions.Citation131 (C) HIV-1 RNA dimerization and circularization. The 5′ UTR PBS region is shown annealed at the PBS to the complementary primer tRNALys3. The dimerization initiation site (DIS) promotes HIV-1 dimerization by forming a “kissing loop” interaction with the DIS of another HIV-1 RNA which transitions to an extended dimer palindromic interface (not shown). Psi, packaging sequence; polyA, polyadenylation signal; and TAR, transactivation response element. U3-R contains complementary sequence to primer tRNALys3.Citation133 Complementary regions in Gag and portions of U3-R are proposed to pair and promote -SSS transfer.Citation132
Acknowledgements
Writing of this review and research in the SBS laboratory was supported by funds from the National Institutes of Health, Public Health Service Grant GM33281 to S.B.S. We thank N. Beliakova-Bethell, M. Zhang and V. Bilanchone, University of California, Irvine for sharing results prior to publication and to colleagues in the retroelement field for helpful discussions over the years.
References
- Eickbush TH, Jamburuthugoda VK. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res 2009; 134:221 - 234
- Symer DE, Boeke JD. Kurth R, Bannert N. An everlasting war dance between retrotransposons and their metazoan hosts. Retroviruses 2010; Norfolk, UK Caister Academic Press 1 - 34
- Beauregard A, Curcio MJ, Belfort M. The take and give between retrotransposable elements and their hosts. Annu Rev Genet 2008; 42:587 - 617
- Bannert N, Fiebig U, Hohn O. Kurth R, Bannert N. Retroviral particles, proteins and genomes. Retroviruses 2010; Norfolk, UK Caister Academic Press 71 - 106
- Engelman A. Kurth R, Bannert N. Reverse transcription and uncoating. Retroviruses 2010; Norfolk, UK Caisert Academic press 129 - 160
- Cimarelli A, Darlix JL. Assembling the human immunodeficiency virus type 1. Cell Mol Life Sci 2002; 59:1166 - 1184
- Bampi C, Jacquenet S, Lener D, Decimo D, Darlix JL. The chaperoning and assistance roles of the HIV-1 nucleocapsid protein in proviral DNA synthesis and maintenance. Int J Biochem Cell Biol 2004; 36:1668 - 1686
- Mougel M, Houzet L, Darlix JL. When is it time for reverse transcription to start and go?. Retrovirology 2009; 6:24
- Thomas JA, Gorelick RJ. Nucleocapsid protein function in early infection processes. Virus Res 2008; 134:39 - 63
- Levin JG, Guo J, Rouzina I, Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog Nucleic Acid Res Mol Biol 2005; 80:217 - 286
- Kurth R, Bannert N. Retroviruses 2010; Norfolk, UK Caister Academic Press
- Ganser-Pornillos BK, Yeager M, Sundquist WI. The structural biology of HIV assembly. Curr Opin Struct Biol 2008; 18:203 - 217
- Scarlata S, Carter C. Role of HIV-1 Gag domains in viral assembly. Biochim Biophys Acta 2003; 1614:62 - 72
- Wilk T, Fuller SD. Towards the structure of the human immunodeficiency virus: divide and conquer. Curr Opin Struct Biol 1999; 9:231 - 243
- Linial ML, Eastman SW. Particle assembly and genome packaging. Curr Top Microbiol Immunol 2003; 277:89 - 110
- Morita E, Sundquist WI. Retrovirus budding. Annu Rev Cell Dev Biol 2004; 20:395 - 425
- Freed EO, Mouland AJ. The cell biology of HIV-1 and other retroviruses. Retrovirology 2006; 3:77
- Yu SF, Sullivan MD, Linial ML. Evidence that the human foamy virus genome is DNA. J Virol 1999; 73:1565 - 1572
- Dvorin JD, Malim MH. Intracellular trafficking of HIV-1 cores: journey to the center of the cell. Curr Top Microbiol Immunol 2003; 281:179 - 208
- Voytas DF, Boeke JD. Craig NL, Craigie R, Gellert M, Lambowitz AM. Ty1 and Ty5 of Saccharomyces cerevisiae. Mobile DNA II 2002; Washington DC American Society for Microbiology 631 - 662
- Sandmeyer SB, Aye M, Menees TM. Ty3: a positionspecific gypsylike element in Saccharomyces cerevisiae 2002; Washington DC ASM Press
- Kim A, Terzian C, Santamaria P, Pelisson A, Purd'homme N, Bucheton A. Retroviruses in invertebrates: the gypsy retrotransposon is apparently an infectious retrovirus of Drosophila melanogaster. Proc Natl Acad Sci USA 1994; 91:1285 - 1289
- Gao X, Rowley DJ, Gai X, Voytas DF. Ty5 gag mutations increase retrotransposition and suggest a role for hydrogen bonding in the function of the nucleocapsid zinc finger. J Virol 2002; 76:3240 - 3247
- Kirchner J, Sandmeyer SB, Forrest DB. Transposition of a Ty3 GAG3-POL3 fusion mutant is limited by availability of capsid protein. J Virol 1992; 66:6081 - 6092
- Farabaugh PJ, Zhao H, Vimaladithan A. A novel programed frameshift expresses the POL3 gene of retrotransposon Ty3 of yeast: frameshifting without tRNA slippage. Cell 1993; 74:93 - 103
- Krausslich HG, Facke M, Heuser AM, Konvalinka J, Zentgraf H. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J Virol 1995; 69:3407 - 3419
- Craven RC, Leure-duPree AE, Erdie CR, Wilson CB, Wills JW. Necessity of the spacer peptide between CA and NC in the Rous sarcoma virus gag protein. J Virol 1993; 67:6246 - 6252
- Zhang M. Ty3 virus-like particle morphogenesis and assembly 2007; Irvine, CA University of California, Irvine
- Kirchner J, Sandmeyer S. Proteolytic processing of Ty3 proteins is required for transposition. J Virol 1993; 67:19 - 28
- Claypool JA, Malik HS, Eickbush TH, Sandmeyer SB. Ten-kilodalton domain in Ty3 Gag3-Pol3p between PR and RT is dispensable for Ty3 transposition. J Virol 2001; 75:1557 - 1560
- Hansen LJ, Chalker DL, Orlinsky KJ, Sandmeyer SB. Ty3 GAG3 and POL3 genes encode the components of intracellular particles. J Virol 1992; 66:1414 - 1424
- Orlinsky KJ, Sandmeyer SB. The Cys-His motif of Ty3 NC can be contributed by Gag3 or Gag3-Pol3 polyproteins. J Virol 1994; 68:4152 - 4166
- Bilanchone VW, Claypool JA, Kinsey PT, Sandmeyer SB. Positive and negative regulatory elements control expression of the yeast retrotransposon Ty3. Genetics 1993; 134:685 - 700
- Kinsey PT, Sandmeyer SB. Ty3 transposes in mating populations of yeast: a novel transposition assay for Ty3. Genetics 1995; 139:81 - 94
- Hansen LJ, Chalker DL, Sandmeyer SB. Ty3, a yeast retrotransposon associated with tRNA genes, has homology to animal retroviruses. Mol Cell Biol 1988; 8:5245 - 5256
- Beliakova-Bethell N, Beckham C, Giddings TH Jr, Winey M, Parker R, Sandmeyer S. Virus-like particles of the Ty3 retrotransposon assemble in association with P-body components. RNA 2006; 12:94 - 101
- Campbell S, Vogt VM. In vitro assembly of virus-like particles with Rous sarcoma virus Gag deletion mutants: identification of the p10 domain as a morphological determinant in the formation of spherical particles. J Virol 1997; 71:4425 - 4435
- Campbell S, Rein A. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J Virol 1999; 73:2270 - 2279
- Zhang Y, Qian H, Love Z, Barklis E. Analysis of the assembly function of the human immunodeficiency virus type 1 gag protein nucleocapsid domain. J Virol 1998; 72:1782 - 1789
- Muriaux D, Mirro J, Harvin D, Rein A. RNA is a structural element in retrovirus particles. Proc Natl Acad Sci USA 2001; 98:5246 - 5251
- Wright ER, Schooler JB, Ding HJ, Kieffer C, Fillmore C, Sundquist WI, et al. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J 2007; 26:2218 - 2226
- Larsen LS, Zhang M, Beliakova-Bethell N, Bilanchone V, Lamsa A, Nagashima K, et al. Ty3 capsid mutations reveal early and late functions of the amino-terminal domain. J Virol 2007; 81:6957 - 6972
- Zhang M, Larsen LS, Irwin B, Bilanchone V, Sandmeyer S. Two-hybrid analysis of Ty3 capsid subdomain interactions. Mob DNA 2010; 1:14
- Larsen LS, Beliakova-Bethell N, Bilanchone V, Zhang M, Lamsa A, Dasilva R, et al. Ty3 nucleocapsid controls localization of particle assembly. J Virol 2008; 82:2501 - 2514
- Kuznetsov YG, Zhang M, Menees TM, McPherson A, Sandmeyer S. Investigation by atomic force microscopy of the structure of Ty3 retrotransposon particles. J Virol 2005; 79:8032 - 8045
- Klein KC, Reed JC, Lingappa JR. Intracellular destinies: degradation, targeting, assembly and endocytosis of HIV Gag. AIDS Rev 2007; 9:150 - 161
- Bieniasz PD. The cell biology of HIV-1 virion genesis. Cell Host Microbe 2009; 5:550 - 558
- Butsch M, Boris-Lawrie K. Destiny of unspliced retroviral RNA: ribosome and/or virion?. J Virol 2002; 76:3089 - 3094
- Cochrane AW, McNally MT, Mouland AJ. The retrovirus RNA trafficking granule: from birth to maturity. Retrovirology 2006; 3:18
- Malim MH, Emerman M. HIV-1 accessory proteins—ensuring viral survival in a hostile environment. Cell Host Microbe 2008; 3:388 - 398
- Nekhai S, Jeang KT. Transcriptional and post-transcriptional regulation of HIV-1 gene expression: role of cellular factors for Tat and Rev. Future Microbiol 2006; 1:417 - 426
- Bray M, Prasad S, Dubay JW, Hunter E, Jeang KT, Rekosh D, et al. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc Natl Acad Sci USA 1994; 91:1256 - 1260
- Levin JG, Rosenak MJ. Synthesis of murine leukemia virus proteins associated with virions assembled in actinomycin D-treated cells: evidence for persistence of viral messenger RNA. Proc Natl Acad Sci USA 1976; 73:1154 - 1158
- Flynn JA, Telesnitsky A. Two distinct Moloney murine leukemia virus RNAs produced from a single locus dimerize at random. Virology 2006; 344:391 - 400
- Butterfield-Gerson KL, Scheifele LZ, Ryan EP, Hopper AK, Parent LJ. Importin-beta family members mediate alpharetrovirus gag nuclear entry via interactions with matrix and nucleocapsid. J Virol 2006; 80:1798 - 1806
- Gudleski N, Flanagan JM, Ryan EP, Bewley MC, Parent LJ. Directionality of nucleocytoplasmic transport of the retroviral gag protein depends on sequential binding of karyopherins and viral RNA. Proc Natl Acad Sci USA 2010; 107:9358 - 9363
- Swanson CM, Puffer BA, Ahmad KM, Doms RW, Malim MH. Retroviral mRNA nuclear export elements regulate protein function and virion assembly. EMBO J 2004; 23:2632 - 2640
- Anderson EC, Lever AM. Human immunodeficiency virus type 1 Gag polyprotein modulates its own translation. J Virol 2006; 80:10478 - 10486
- Berkowitz RD, Ohagen A, Hoglund S, Goff SP. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J Virol 1995; 69:6445 - 6456
- Zhang Y, Barklis E. Nucleocapsid protein effects on the specificity of retrovirus RNA encapsidation. J Virol 1995; 69:5716 - 5722
- D'Souza V, Summers MF. How retroviruses select their genomes. Nat Rev Microbiol 2005; 3:643 - 655
- D'Souza V, Summers MF. Structural basis for packaging the dimeric genome of Moloney murine leukaemia virus. Nature 2004; 431:586 - 590
- De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science 1998; 279:384 - 388
- Johnson MC, Scobie HM, Ma YM, Vogt VM. Nucleic acid-independent retrovirus assembly can be driven by dimerization. J Virol 2002; 76:11177 - 11185
- Larson DR, Ma YM, Vogt VM, Webb WW. Direct measurement of Gag-Gag interaction during retrovirus assembly with FRET and fluorescence correlation spectroscopy. J Cell Biol 2003; 162:1233 - 1244
- Jouvenet N, Bieniasz PD, Simon SM. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature 2008; 454:236 - 240
- Perlman M, Resh MD. Identification of an intracellular trafficking and assembly pathway for HIV-1 gag. Traffic 2006; 7:731 - 745
- Resh MD. Intracellular trafficking of HIV-1 Gag: how Gag interacts with cell membranes and makes viral particles. AIDS Rev 2005; 7:84 - 91
- Dooher JE, Schneider BL, Reed JC, Lingappa JR. Host ABCE1 is at plasma membrane HIV assembly sites and its dissociation from Gag is linked to subsequent events of virus production. Traffic 2007; 8:195 - 211
- Basyuk E, Boulon S, Skou Pedersen F, Bertrand E, Vestergaard Rasmussen S. The packaging signal of MLV is an integrated module that mediates intracellular transport of genomic RNAs. J Mol Biol 2005; 354:330 - 339
- Poole E, Strappe P, Mok HP, Hicks R, Lever AM. HIV-1 Gag-RNA interaction occurs at a perinuclear/centrosomal site; analysis by confocal microscopy and FRET. Traffic 2005; 6:741 - 755
- Grigorov B, Decimo D, Smagulova F, Pechoux C, Mougel M, Muriaux D, et al. Intracellular HIV-1 Gag localization is impaired by mutations in the nucleocapsid zinc fingers. Retrovirology 2007; 4:54
- Levesque K, Halvorsen M, Abrahamyan L, Chatel-Chaix L, Poupon V, Gordon H, et al. Trafficking of HIV-1 RNA is mediated by heterogeneous nuclear ribonucleoprotein A2 expression and impacts on viral assembly. Traffic 2006; 7:1177 - 1193
- Chen C, Jin J, Rubin M, Huang L, Sturgeon T, Weixel KM, et al. Association of gag multimers with filamentous actin during equine infectious anemia virus assembly. Curr HIV Res 2007; 5:315 - 323
- Krogstad P, Geng YZ, Rey O, Canon J, Ibarrondo FJ, Ackerson B, et al. Human immunodeficiency virus nucleocapsid protein polymorphisms modulate the infectivity of RNA packaging mutants. Virology 2002; 294:282 - 288
- Liu B, Dai R, Tian CJ, Dawson L, Gorelick R, Yu XF. Interaction of the human immunodeficiency virus type 1 nucleocapsid with actin. J Virol 1999; 73:2901 - 2908
- Wilk T, Gowen B, Fuller SD. Actin associates with the nucleocapsid domain of the human immunodeficiency virus Gag polyprotein. J Virol 1999; 73:1931 - 1940
- Hong S, Choi G, Park S, Chung AS, Hunter E, Rhee SS. Type D retrovirus Gag polyprotein interacts with the cytosolic chaperonin TRiC. J Virol 2001; 75:2526 - 2534
- Lingappa JR, Dooher JE, Newman MA, Kiser PK, Klein KC. Basic residues in the nucleocapsid domain of Gag are required for interaction of HIV-1 gag with ABCE1 (HP68), a cellular protein important for HIV-1 capsid assembly. J Biol Chem 2006; 281:3773 - 3784
- Malagon F, Jensen TH. The T body, a new cytoplasmic RNA granule in Saccharomyces cerevisiae. Mol Cell Biol 2008; 28:6022 - 6032
- Dutko JA, Kenny AE, Gamache ER, Curcio MJ. 5′ to 3′ mRNA decay factors colocalize with Ty1 gag and human APOBEC3G and promote Ty1 retrotransposition. J Virol 2010; 84:5052 - 5066
- Checkley MA, Nagashima K, Lockett SJ, Nyswaner KM, Garfinkel DJ. P-body components are required for Ty1 retrotransposition during assembly of retrotransposition-competent virus-like particles. Mol Cell Biol 2010; 30:382 - 398
- Griffith JL, Coleman LE, Raymond AS, Goodson SG, Pittard WS, Tsui C, et al. Functional genomics reveals relationships between the retrovirus-like Ty1 element and its host Saccharomyces cerevisiae. Genetics 2003; 164:867 - 879
- Irwin B, Aye M, Baldi P, Beliakova-Bethell N, Cheng H, Dou Y, et al. Retroviruses and yeast retrotransposons use overlapping sets of host genes. Genome Res 2005; 15:641 - 654
- Balagopal V, Parker R. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr Opin Cell Biol 2009; 21:403 - 408
- Nissan T, Parker R. Analyzing P-bodies in Saccharomyces cerevisiae. Methods Enzymol 2008; 448:507 - 520
- Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell 2009; 36:932 - 941
- Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol 2008; 183:441 - 455
- Hu W, Sweet TJ, Chamnongpol S, Baker KE, Coller J. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature 2009; 461:225 - 229
- Hoyle NP, Castelli LM, Campbell SG, Holmes LE, Ashe MP. Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J Cell Biol 2007; 179:65 - 74
- Kedersha N, Anderson P. Mammalian stress granules and processing bodies. Methods Enzymol 2007; 431:61 - 81
- Kapp LD, Lorsch JR. The molecular mechanics of eukaryotic translation. Annu Rev Biochem 2004; 73:657 - 704
- Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell 2005; 122:875 - 886
- Coller JM, Tucker M, Sheth U, Valencia-Sanchez MA, Parker R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA 2001; 7:1717 - 1727
- Gorelick RJ, Henderson LE, Hanser JP, Rein A. Point mutants of Moloney murine leukemia virus that fail to package viral RNA: evidence for specific RNA recognition by a “zinc finger-like” protein sequence. Proc Natl Acad Sci USA 1988; 85:8420 - 8424
- Dupraz P, Oertle S, Meric C, Damay P, Spahr PF. Point mutations in the proximal Cys-His box of Rous sarcoma virus nucleocapsid protein. J Virol 1990; 64:4978 - 4987
- Dorfman T, Luban J, Goff SP, Haseltine WA, Gottlinger HG. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J Virol 1993; 67:6159 - 6169
- Beliakova-Bethell N, Terry LJ, Bilanchone V, DaSilva R, Nagashima K, Wente SR, et al. Ty3 nuclear entry is initiated by viruslike particle docking on GLFG nucleoporins. J Virol 2009; 83:11914 - 11925
- Scheifele LZ, Garbitt RA, Rhoads JD, Parent LJ. Nuclear entry and CRM1-dependent nuclear export of the Rous sarcoma virus Gag polyprotein. Proc Natl Acad Sci USA 2002; 99:3944 - 3949
- Garbitt-Hirst R, Kenney SP, Parent LJ. Genetic evidence for a connection between Rous sarcoma virus gag nuclear trafficking and genomic RNA packaging. J Virol 2009; 83:6790 - 6797
- Rey O, Canon J, Krogstad P. HIV-1 Gag protein associates with F-actin present in microfilaments. Virology 1996; 220:530 - 534
- Gladnikoff M, Shimoni E, Gov NS, Rousso I. Retroviral assembly and budding occur through an actin-driven mechanism. Biophys J 2009; 97:2419 - 2428
- Mouland AJ, Mercier J, Luo M, Bernier L, DesGroseillers L, Cohen EA. The double-stranded RNA-binding protein Staufen is incorporated in human immunodeficiency virus type 1: evidence for a role in genomic RNA encapsidation. J Virol 2000; 74:5441 - 5451
- Chatel-Chaix L, Boulay K, Mouland AJ, Desgroseillers L. The host protein Staufen1 interacts with the Pr55Gag zinc fingers and regulates HIV-1 assembly via its N-terminus. Retrovirology 2008; 5:41
- Bacharach E, Gonsky J, Alin K, Orlova M, Goff SP. The carboxy-terminal fragment of nucleolin interacts with the nucleocapsid domain of retroviral gag proteins and inhibits virion assembly. J Virol 2000; 74:11027 - 11039
- Popov S, Popova E, Inoue M, Gottlinger HG. Human immunodeficiency virus type 1 Gag engages the Bro1 domain of ALIX/AIP1 through the nucleocapsid. J Virol 2008; 82:1389 - 1398
- Dussupt V, Javid MP, Abou-Jaoude G, Jadwin JA, de La Cruz J, Nagashima K, et al. The nucleocapsid region of HIV-1 Gag cooperates with the PTAP and LYPXnL late domains to recruit the cellular machinery necessary for viral budding. PLoS Pathog 2009; 5:1000339
- Popova E, Popov S, Gottlinger HG. Human immunodeficiency virus type 1 nucleocapsid p1 confers ESCRT pathway dependence. J Virol 2010; 84:6590 - 6597
- Luo K, Liu B, Xiao Z, Yu Y, Yu X, Gorelick R, et al. Amino-terminal region of the human immunodeficiency virus type 1 nucleocapsid is required for human APOBEC3G packaging. J Virol 2004; 78:11841 - 11852
- Alce TM, Popik W. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J Biol Chem 2004; 279:34083 - 34086
- Schafer A, Bogerd HP, Cullen BR. Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the gag polyprotein precursor. Virology 2004; 328:163 - 168
- Barbee SA, Estes PS, Cziko AM, Hillebrand J, Luedeman RA, Coller JM, et al. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron 2006; 52:997 - 1009
- Furtak V, Mulky A, Rawlings SA, Kozhaya L, Lee K, Kewalramani VN, et al. Perturbation of the P-body component Mov10 inhibits HIV-1 infectivity. PLoS One 2010; 5:9081
- Gallois-Montbrun S, Kramer B, Swanson CM, Byers H, Lynham S, Ward M, et al. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J Virol 2007; 81:2165 - 2178
- Nathans R, Chu CY, Serquina AK, Lu CC, Cao H, Rana TM. Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol Cell 2009; 34:696 - 709
- Chable-Bessia C, Meziane O, Latreille D, Triboulet R, Zamborlini A, Wagschal A, et al. Suppression of HIV-1 replication by microRNA effectors. Retrovirology 2009; 6:26
- Pettit SC, Moody MD, Wehbie RS, Kaplan AH, Nantermet PV, Klein CA, et al. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J Virol 1994; 68:8017 - 8027
- Qualley DF, Stewart-Maynard KM, Wang F, Mitra M, Gorelick RJ, Rouzina I, et al. C-terminal domain modulates the nucleic acid chaperone activity of human T-cell leukemia virus type 1 nucleocapsid protein via an electrostatic mechanism. J Biol Chem 2010; 285:295 - 307
- Stewart-Maynard KM, Cruceanu M, Wang F, Vo MN, Gorelick RJ, Williams MC, et al. Retroviral nucleocapsid proteins display nonequivalent levels of nucleic acid chaperone activity. J Virol 2008; 82:10129 - 10142
- Cruceanu M, Urbaneja MA, Hixson CV, Johnson DG, Datta SA, Fivash MJ, et al. Nucleic acid binding and chaperone properties of HIV-1 Gag and nucleocapsid proteins. Nucleic Acids Res 2006; 34:593 - 605
- Chapman KB, Bystrom AS, Boeke JD. Initiator methionine tRNA is essential for Ty1 transposition in yeast. Proc Natl Acad Sci USA 1992; 89:3236 - 3240
- Keeney JB, Chapman KB, Lauermann V, Voytas DF, Astrom SU, von Pawel-Rammingen U, et al. Multiple molecular determinants for retrotransposition in a primer tRNA. Mol Cell Biol 1995; 15:217 - 226
- Ke N, Gao X, Keeney JB, Boeke JD, Voytas DF. The yeast retrotransposon Ty5 uses the anticodon stemloop of the initiator methionine tRNA as a primer for reverse transcription. RNA 1999; 5:929 - 938
- Lin JH, Levin HL. A complex structure in the mRNA of Tf1 is recognized and cleaved to generate the primer of reverse transcription. Genes Dev 1997; 11:270 - 285
- Cristofari G, Ficheux D, Darlix JL. The GAG-like protein of the yeast Ty1 retrotransposon contains a nucleic acid chaperone domain analogous to retroviral nucleocapsid proteins. J Biol Chem 2000; 275:19210 - 19217
- Clark DJ, Bilanchone VW, Haywood LJ, Dildine SL, Sandmeyer SB. A yeast sigma composite element, TY3, has properties of a retrotransposon. J Biol Chem 1988; 263:1413 - 1423
- Gabus C, Ficheux D, Rau M, Keith G, Sandmeyer S, Darlix JL. The yeast Ty3 retrotransposon contains a 5′-3′ bipartite primer-binding site and encodes nucleocapsid protein NCp9 functionally homologous to HIV-1 NCp7. EMBO J 1998; 17:4873 - 4880
- Cristofari G, Gabus C, Ficheux D, Bona M, Le Grice SF, Darlix JL. Characterization of active reverse transcriptase and nucleoprotein complexes of the yeast retrotransposon Ty3 in vitro. J Biol Chem 1999; 274:36643 - 36648
- Friant S, Heyman T, Bystrom AS, Wilhelm M, Wilhelm FX. Interactions between Ty1 retrotransposon RNA and the T and D regions of the tRNA(iMet) primer are required for initiation of reverse transcription in vivo. Mol Cell Biol 1998; 18:799 - 806
- Friant S, Heyman T, Wilhelm ML, Wilhelm FX. Extended interactions between the primer tRNAi(Met) and genomic RNA of the yeast Ty1 retrotransposon. Nucleic Acids Res 1996; 24:441 - 449
- Cristofari G, Bampi C, Wilhelm M, Wilhelm FX, Darlix JL. A 5′-3′ long-range interaction in Ty1 RNA controls its reverse transcription and retrotransposition. EMBO J 2002; 21:4368 - 4379
- Ooms M, Abbink TE, Pham C, Berkhout B. Circularization of the HIV-1 RNA genome. Nucleic Acids Res 2007; 35:5253 - 5261
- Piekna-Przybylska D, DiChiacchio L, Mathews DH, Bambara RA. A sequence similar to tRNA 3 Lys gene is embedded in HIV-1 U3-R and promotes minus-strand transfer. Nat Struct Mol Biol 2010; 17:83 - 89
- Song M, Balakrishnan M, Gorelick RJ, Bambara RA. A succession of mechanisms stimulate efficient reconstituted HIV-1 minus strand strong stop DNA transfer. Biochemistry 2009; 48:1810 - 1819
- Chapman KB, Boeke JD. Isolation and characterization of the gene encoding yeast debranching enzyme. Cell 1991; 65:483 - 492
- Bushman FD, Malani N, Fernandes J, D'Orso I, Cagney G, Diamond TL, et al. Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathog 2009; 5:1000437
- Cheng Z, Menees TM. RNA branching and debranching in the yeast retrovirus-like element Ty1. Science 2004; 303:240 - 243
- Ye Y, De Leon J, Yokoyama N, Naidu Y, Camerini D. DBR1 siRNA inhibition of HIV-1 replication. Retrovirology 2005; 2:63
- Coombes CE, Boeke JD. An evaluation of detection methods for large lariat RNAs. RNA 2005; 11:323 - 331
- Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW Jr, Swanstrom R, et al. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 2009; 460:711 - 716