Abstract
Claudins form the paracellular tight junction seal in epithelial tissues. Although there is still limited information on how these proteins are organized at the junction, a number of recent studies have provided useful insights both into claudin-claudin interactions and into interactions between claudins and other proteins. The focus of this review is to summarize recent information about claudin interactions and to identify critical unanswered questions about claudin organization and tight junction structure which will be required to understand claudin function.
Introduction
Half a century ago, Farquhar and Palade described the ultrastructure of the apical junctional complex of epithelial cells.Citation1 They divided this region into three neighboring structures: the apical zonula occludens or tight junction, the adherens junction, just below the tight junction, and the macula adherens or desmosomes that form spot-like contacts on the lateral membranes. In transmission electron microscopy, the tight junction was distinguished by close apposition of the plasma membranes and several spots, or “kisses,” where the membranes of the adjacent cells appear to fuse. Freeze-fracture electron microscopy of the same region revealed a series of strands that encircle the apical end of the lateral membraneCitation2; these strands correspond to the regions of membrane contact detected in transmission electron microscopy and represent a series of barriers to the diffusion of material across the epithelium between adjacent cells. Of note, when tissues are fractured without prior fixation with aldehyde the strands appear as rows of discrete 10nm particles.Citation3 It is tempting to speculate that the particles represent a basic structural unit of claudin interacting proteins although there is currently no biochemical evidence to support this notion. In 1998, Furuse and Tsukita demonstrated that expression of claudins, small four-TM proteins they had recently localized to the tight junction, could recapitulate similar freeze fracture strands when expressed in non-epithelial cells.Citation4 They and others have subsequently clearly demonstrated that claudins are the critical sealing proteins of the tight junction. Variable tissue-specific expression of different combinations of the 23+ membersCitation5 of the claudin family of proteins is likely to explain the observed physiologic differences in the sealing properties of tight junctions in different epithelia.Citation6 Much progress has been made in understanding the contributions of different claudins to barrier functions. In addition, many other proteins have been localized to tight junctions and several of these are thought to interact with claudins directly.
In spite of our insights into the protein components of the barrier, we still lack a detailed understanding of the molecular structure and protein interactions of tight junctions. However, below we review a number of recent studies that better define claudin interactions and provide some organizational clues. Both microscopic and biochemical techniques have allowed identification of critical regions in transcellular claudin-claudin interactions. Physiologic and genetic analyses of claudin mutants have suggested regions important in the sealing properties of claudins and thus offer hints about the structural organization of their extracellular domains. Other studies have demonstrated the requirement for heterologous oligomerization of some claudins for tight junction targeting, which may lead to identification of domains involved in both localization at the barrier and into regions required for specific claudin-claudin interactions.
Several recent studies have also examined the interactions of claudins with other tight junction proteins, including occludin and other members of the Marvel-domain containing family. In addition, along with the initial recognition of claudin interactions with the tight junction scaffolding proteins, ZO-1, -2 and -3, there have been several studies on the interaction of claudins with other PDZ-domain containing proteins. Finally, some claudins have been shown to interact with non-tight junction proteins, including the cell adhesion proteins EpCam and tetraspanins and the signaling proteins, ephrin A and B and their receptors, EphA and EphB. The role of these and other proteins interacting with lateral membrane claudins is likely to provide important information about how the non-tight junction, non-barrier, pool of claudin is organized and regulated.
Finally, several recent papers that better define the ultrastructural anatomy of the tight junction suggest that the overall organization of junctions is likely to be more complex than commonly thought, with differences in protein composition and structure in different subdomains of a single tight junction.Citation7,Citation8 All of this new information is helpful in understanding the tight junction, but better molecular models require structural information of the component proteins and better understanding of protein-protein interactions, which are still lacking.
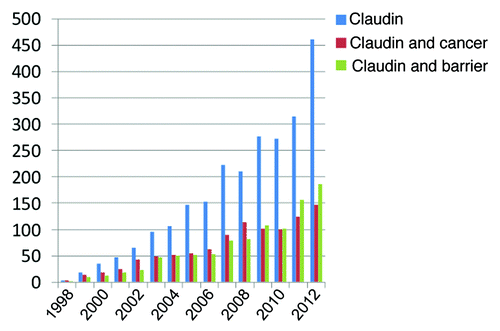
Although the focus of this review and this journal is on epithelia barriers it is worth noting that there has been significant interest in the role of claudins in cancer. displays the number of publications in PubMed each year since claudins were first described in 1998 by searching with MeSH terms “Claudin,” “Claudin and cancer” or “Claudin and barrier.” Overall publications on claudins continue to grow at a nonlinear rate and with approximately equal focus on their role in cancer and in forming the tight junction barrier. In this review we provide a survey of protein-protein interactions which should inform the function of claudins in both cancer and in forming the paracellular barrier. We focus on cases where there is published evidence of a direct protein interaction with a claudin and acknowledge there is suggestive evidence for indirect interactions with more.
Claudin-Claudin interactions
As mentioned above, soon after claudins were identified as tight junction constituents, Furuse et al.Citation4 demonstrated that expression of either claudin-1 or claudin-2 in fibroblasts resulted in the formation of continuous linear polymeric rows that were similar to the fibrils seen in cultured epithelial cells and epithelial tissues by freeze fracture electron microscopy. Since then, exogenous expression of many different claudins in either fibroblasts or cultured epithelial cells has been demonstrated to result in tight junction-like fibrils.Citation9-Citation15 In biochemical assays, claudins have been demonstrated to form multimers, but the structure and subunit number of polymerized claudins is unclear.Citation16,Citation17 To better define the ability of individual claudins to polymerize into freeze fracture fibrils, Yamazaki et al.Citation18 expressed several different claudins in an epithelial-like cell line, S7 cells, which lacks most claudins and does not normally contain tight junction strands. These authors found that in these cells, claudin-7,-14, and -19, but not most other claudins, could polymerize and form strands when stably expressed. Several other transfected claudins were distributed on the lateral membrane, but did not polymerize to form strands. Since some of the same claudins (claudins 1, 2, 3, and 10 as examples) that were studied by Yamazaki et al. can form strands in other non-epithelial cells,Citation4,Citation19 it appears that the polymerization of claudins may be affected by the cell background, perhaps requiring cell-specific post-translational modification or interaction with a cell-specific protein(s). In this context, it is notable that although in the study by Yamazaki et al., claudin-7 had among the strongest polymerization propensity, in many epithelial cells, claudin-7 is localized not at the tight junction, but on the lateral call membrane, without associated lateral strands.Citation20 Together, these results support the notion that although claudins likely oligomerize to form the tight junction strands, this process can be influenced by other cellular components.
One of the most dramatic pieces of evidence supporting the role of other cellular proteins in the organization of the freeze fracture strands was also demonstrated by Ikenouchi et al.Citation21 In this study, expression of claudin-1 in L cell fibroblasts resulted in linear, non-branching strand arrays, while co-expression of tricellulin with claudin-1 resulted in a highly branched network of strands. Expression of tricellulin alone does not result in strand formation. The morphology of freeze fracture stands varies widely among epithelia and it seems likely that morphology is regulated by the profile of claudins as well as interacting proteins expressed in each cell type.
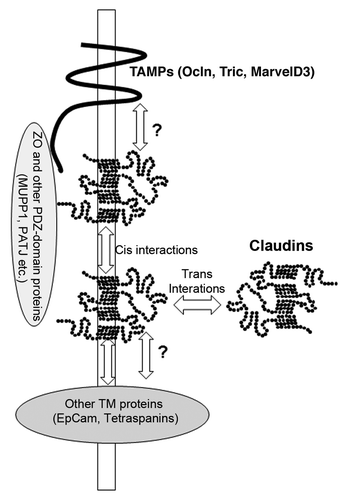
In epithelial cells and tissues, it appears that most tight junctions contain more than one claudin type. Several studies have demonstrated that multiple claudins are present in a single tight junction strand, suggesting that these different claudin gene products can co-polymerize. However, the site and mechanisms of this side to side (cis) interaction within the same cell are unclear (these and other interactions are presented schematically in ). Recent studies by Piontek et al.Citation22 have begun to catalog the ability of different claudins to form heteropolymers. These authors have focused on those claudins that are expressed in cerebral vasculature, including claudin 1, 2, 3, 5, and 12. Fluorescently-tagged claudins were transfected pair-wise into HEK293 cells and interactions detected by fluorescence resonance energy transfer (FRET). They found that most claudins that they studied (but not claudin-12) could interact in cis, although not equally well. The distance between the CFP and YFP tags used for the assay was less than 7 nm,Citation23 which does not prove direct interaction, but is consistent with close proximity.
Figure 2. Model of claudin interactions with other proteins. The question marks refer to the possibility that interactions between claudins and TAMPs and claudins and tetraspanins may in part be mediated by the extracellular domains (see text).
Two special cases of cis heterotypic claudin interactions have recently been identified by Hou et al.Citation24,Citation25 These very interesting studies demonstrated a requirement for cis interactions between claudin-16 and claudin-19 and between claudin-4 and claudin-8 for proper targeting. Exogenously expressed claudin-19 is localized to tight junctions but claudin-16 is not. However, when co-expressed with claudin-19, claudin-16 was efficiently incorporated into tight junction strands. Direct interaction between these two proteins (and in a similar study, between claudin-4 and claudin-8) was demonstrated by yeast-2-hybrid analysis; the results suggested that interactions between the different claudins might be mediated by their transmembrane domains and that this is required to recruit or guide some claudins to the tight junction.
Claudins associate not only in cis, but also across the paracellular space, in trans. Early studies demonstrated that claudin-1, -2, and -3 could interact homotypically in trans, and that only claudin-1 and -3 but not -2 could interact heterotypically with claudins on adjacent cells.Citation15 Both extracellular domains have been reported to contribute to this binding interaction.Citation26-Citation29 In spite of considerable sequence conservation among different claudins, heterotypic trans interactions between different claudins appear to be relatively rare. For example, claudin-3 and -4 share considerable sequence identity in the first extracellular domain, but do not interact in trans. However, mutation of a single amino acid in the first extracellular domain of claudin-3 was sufficient to promote interaction with claudin-4.Citation29 Similarly, mutational analysis of a single critical tyrosine residue in the second extracellular domain can prohibit interactions between claudins -1 and -5.Citation22 Overall, the published data suggest that the sequence requirements for trans interactions across the cells are much stricter than cis interactions in the strands of a single bilayer.
Although in vitro studies suggest that the trans interactions for individual claudins are weak,Citation28 several pieces of evidence suggest that polymerized claudins are likely to form very stable interactions. First, exogenous expression of claudins in fibroblasts resulted in increased calcium independent cell-cell adhesion.Citation30 Second, when cells singly expressing claudins labeled with different fluorescent tags were co-cultured, microscopic analysis of endocytosed claudins revealed vesicles contained both fluorescent tags, suggesting that the tight junction containing membranes from the apposed cells were co-endocytosed, referred to by the authors as “eat each other endocytosis”Citation31. Finally, FRAP analysis of claudins in epithelial cells revealed a very low level of mobility, consistent with the ability of these proteins to form stable polymers with low diffusion within the membrane.Citation32 However, it is worth noting that when fluorescent (GFP-fused) claudins were expressed in fibroblasts, the paired strands formed between cells were dynamic in that they broke and reformed continuously.Citation33 Although this dynamic behavior has not been visualized in epithelial cells, it is possible that it occurs at a slower time frame. This behavior could allow the tight junction seal to be maintained as adjacent epithelial cells shift position relative to each other or divide and apoptose.
Claudin-Occludin/Tricellulin/Marvel D3 Interactions
Occludin was much heralded as the first identified tight junction transmembrane protein,Citation34 but occludin gene deletion was subsequently shown to have little effect on the paracellular barrier or on tight junction strand formation.Citation35 Although exogenous expression of occludin does not result in tight junction strand formation, occludin is recruited to the tight junction strands when co-expressed with claudins.Citation4 Cording et al.Citation36 have recently extended these observations and used a combination of techniques to study interactions between tight junction members of the occludin family (TAMPs, for tight junction-associated Marvel domain proteins) occludin, tricellulinCitation37 and Marvel D3.Citation38 Cording and coworkers demonstrated that expression of either claudin-1 or claudin-5 in HEK293 cells resulted in increased recruitment of tagged occludin, tricellulin and Marvel D3 to sites of cell contact as measured by FRET. FRAP analysis demonstrated that co-expression with claudin-1 stabilized TAMPs at the sites of cell-cell contact. Additionally, this group found that co-expression of claudin-1 with any of the TAMPs resulted in differing freeze fracture strand patterns. Co-expression of claudin-1 with occludin or Marvel D3 increased the number of parallel freeze fracture strands, while Marvel D3/claudin-1 co-expression also increased the overall complexity of cross-bridges among tight junction strands. In contrast, co-expression of tricellulin with claudin-1 dramatically and specifically increased the cross-bridging of the strands. This group noted that there were differences in the strength of interactions between different members of the claudin and TAMP families. For example, the interaction between occludin and claudin-1 was stronger than the interaction between tricellulin and claudin-1. Reasoning that more stable claudin-1/occludin interaction might dominate over the tricellulin/claudin-1 interaction, they knocked down claudin-1 in Caco-2 cells and noted redistribution of tricellulin to bicellular border. They suggested that higher affinity interactions between claudin-1 and occludin than between claudin-1 and tricellulin normally result in the exclusion of tricellulin from bicellular contacts and thus may in part explain its restriction to tricellular cell contacts.
The nature of the interaction of claudin and occludin (or other TAMPs) is not well understood. Both proteins can separately interact with the tight junction scaffolding protein ZO-1 (described in more detail below). Raleigh and coworkersCitation39 showed a ZO-1 dependent interaction between the tail of occludin and claudin-1 and -2; this interaction was influenced by the level of occludin phosphorylation at serine 408. However, ZO-1 binding is not required for occludin localization, since occludin with the ZO-1 binding domain deleted is recruited to cell contacts in claudin-expressing fibroblasts but not untransfected cells (our unpublished results). In addition, in the above studies,Citation36 the claudins being investigated had fluorescent protein tags at their C-termini, blocking the PDZ-binding motif required for ZO-1 interaction but were still able to recruit TAMPs to cell contacts. Data published by Harris and coworkersCitation40 demonstrate that claudin-1-occludin interactions, as measured by FRET analysis can be abrogated by mutational changes at residues I32M or E48K in the first extracellular domain of claudin, suggesting that this region is important for interaction. Interestingly, they also showed that claudin-7, which they find does not normally associate with occludin, shows significant interaction when the corresponding positions in the first loop are mutated to the same residues, M32I, K48E found in wild-type claudin-1. These results support those published earlier by Mrsny and coworkers, who also suggested that claudin and occludin may interact via their extracellular domains; they found a claudin mimetic peptide to a region of the first extracellular domain could be cross-linked to both claudin-1 and occludin.Citation41 They interpreted this result to suggest that the claudin peptide bound to occludin and represents the natural contact region between on claudin for occludin.
The physiologic importance of interactions between occludin and claudins is not clear, since as mentioned above, occludin knockout animals do not show an appreciable barrier defect and have normal appearing tight junctions. However, there are data that suggest that occludin may be important for signaling at the tight junction.Citation39,Citation42,Citation43 One recent report demonstrates that occludin is required for apoptosis following disruption of claudin-claudin interactions.Citation44 In this study, disruption of claudin-4 interactions with a mimic peptide to the second extracellular domain stimulated caspase activation and apoptosis, but not in occludin deficient cells. These authors suggest that occludin may be acting to recruit FADD (Fas-associated protein with death domain) and caspase to a tight junction signaling domain. It seems likely that even if occludin is not required for tight junction formation, that occludin-claudin interactions are likely to be important in the regulated tight junction behavior.
Claudin-ZO Family Interactions
The three ZO scaffolding proteins of the tight junction each contain three PDZ-binding motifs in their N-terminal half (reviewed in ref. Citation45) and almost all claudins (claudin-12 being the exception) have a PDZ-binding motif at the their carboxyl termini.Citation46 Tsukita and colleagues demonstratedCitation47 direct binding of GST- fusion proteins representing the carboxyl tails of claudins 1–8 to His-tagged PDZ-1 of ZO-1, ZO-2, and ZO-3. This PDZ-dependent interaction with ZO proteins is thought to promote proper targeting of claudins to the tight junctionCitation12 and studies with ZO-1/ZO-2 knockdown cells show disruptions in claudin localization and in barrier function.Citation48,Citation49 However, in some cases, in the presence of pre-formed tight junctions, claudins do not require PDZ-dependent interactions with ZO-1 for localization,Citation50,Citation51 presumably due to their ability to oligomerize at the tight junction with non-truncated resident claudins. In contrast, a disease-associated human mutation that disrupts the ability of claudin-16 to interact with ZO-1 resulted in mistargeting to lysosomes and a renal transport defect manifest as childhood hypercalcuria,Citation52 suggesting that the importance of the PDZ interactions may be context-dependent.
Although almost all claudins terminate in the amino acids YV, the other amino acids within the PDZ-binding motif (~6 amino acid residues) that are likely to influence PDZ binding interactions are not well conserved. Comparison of the crystal structure of the first PDZ domain of ZO-1 and that of the adherens junction protein, ErbinCitation53 with synthetic peptide ligands suggests lower specificity for interaction with ZO-1 than for Erbin. This might explain the ability of the first PDZ of ZO-1 to interact with other proteins (adherens junction proteins, for example) along with the variety of claudins.
The PDZ binding motifs of claudins can and do interact with other PDZ domain proteins. Claudin-5, claudin-8, and claudin-4 can all interact with the multi-PDZ domain containing protein MUPP1.Citation54-Citation56 Both MUPP1 and claudin-4 are upregulated by hypertonic stress and can be co-immunoprecipitated from an immortalized collecting duct kidney cell line. Claudin-1 colocalizes with the PDZ-domain containing protein PATJ in the paranodal loops of myelinating Schwann cellsCitation56 and can interact with the 8th PDZ domain of PATJ in epithelial cells.Citation57 Given the large number of PDZ-domain containing proteins at the tight junction and lateral membrane, it seems likely that other interactions between claudins and some of these proteins can occurs as well. However, except for ZO-1, the relative importance of interactions of claudins and other PDZ domain-containing proteins remains to be determined.
Interactions with Other Proteins
EpCAM
Epithelial cell adhesion molecule (EpCAM) is a glycosylated single pass transmembrane protein found in epithelial cells where it forms homophilic calcium-independent bonds with partners on adjacent cells in the lateral plasma membrane. It is also frequently upregulated in cancer and recently implicated in stem cell signaling, reviewed in.Citation58 Claudin-7, which is most often localized to the lateral membrane of epithelial cells, can be co-immunoprecipitated with EpCAM after crosslinking with a membrane-permeant crosslinking agent.Citation59 The EpCAM-claudin-7 complex is concentrated in glycolipid-enriched membranes; this domain is also relatively enriched in phosphorylated claudin-7, suggesting that phosphorylation state might influence its interaction with EpCAM.Citation59 Recent data suggest that the EpCAM-claudin-7 complex is also associated with tetraspanin 8 (CO-029) and a form of CD44,Citation60 although the principal interaction appears to be between EpCAM and claudin-7. Detailed analysis of the interaction of these two proteins demonstrated a critical sequence in the transmembrane domain of EpCAM, and that the cytoplasmic domains of both proteins were dispensable for the interaction.Citation61 This group further showed that interaction with claudin-7 recruited EpCAM into tetraspanin-enriched membrane domains and that not EpCAM alone, but the EpCAM-claudin-7 complex strongly promoted tumorigenicity and accelerated tumor growth.Citation61
EpCAM knockout mice show defects in intestinal barrier function, decreases in the levels of claudin-2, -3 and -15 and loss of claudin-7 along with disarranged tight junction fibrils.Citation62 In contrast, knockdown of EpCAM in intestinal cell lines promotes tight junction formation and results in a remarkable relocalization of claudin-7 from the lateral membrane to the tight junction.Citation63 Although TER and barrier function was increased and claudin-7 was now associated with the tight junction in the EpCAM knockdown cells, Wu and coworkers found a decrease in the levels of claudin-7. This was found to be due to increased degradation and further experiments demonstrated that EpCAM acted to stabilize claudin-7. Although these two studies show differing effects of reducing EpCAM levels on the tight junction, the importance of this EpCAM in claudin-7 localization on the lateral membrane and stability is consistent between the in vivo and in vitro analyses.
Tetraspanins
Tetraspanins are small integral 4TM membrane proteins with a similar topology but no sequence identity to the claudin family of proteins. Tetraspanins are postulated to organize a network of molecular interactions at the cell surface, referred to as a tetraspanin web, including integrins and other adhesive proteins, proteins with immunoglobulin domains, proteases and signaling molecules (reviewed in ref. Citation64). Although the interaction among claudin-7, EpCAM and tetraspanins described above is indirect, there are several reports of direct claudin-tetraspanin interactions. For example, OAP-1 forms a complex with claudin-11 and integrinsCitation65 and chemical crosslinking reveals interaction between claudin-1 and the tetraspanin CD9.Citation66 However, by far the most attention has been focused on the interaction between CD81 and claudin-1, which along with several other proteins, are required for hepatitis C virus infectivity (reviewed in ref. Citation67). This interaction has been most carefully described in a recent study by Harris et al.Citation40 where using FRET analysis, they found mutation of two critical residues in the claudin-1 extracellular domains inhibited both CD81 interaction and binding of hepatitis C virus. In addition, they found that only claudin-1 at the basolateral surface of HepG2 cells was associated with CD81, not the tight junction pool of claudin-1, consistent with basolateral but not tight junction localization for virus infection.
Ephrins
The Eph receptors and their transmembrane ligands, the ephrins, regulate a number of cell-cell signaling pathways in epithelia, brain and the vascular system. Engagement of Ephrin B1 with the Eph receptor kinase on an adjacent cell can result in both downstream signal transduction on cells expressing Ephrin B1 and reverse signaling back through the Eph receptor kinase (reviewed in ref. Citation68). Ephrin B1 has been found to co-immunoprecipitate with and partially colocalize with claudin-1 and claudin-4 in both HT29 and MDCK cells; this interaction was demonstrated to depend on the extracellular domains of both proteins.Citation69 The cytoplasmic domain of Ephrin B1 is phosphorylated upon the formation of cell-cell contact and phosphorylation was shown to be dependent on the presence of the claudin extracellular domain identified as critical in the interaction between these two proteins. Overexpression of Ephrin B1 in MDCK cells results in small but significant changes in barrier function,Citation69 suggesting that this interaction may be a component of normal tight junction regulation. Evidence also points to a role for interaction between claudins and Ephrin B during normal development, since both claudin-1 and claudin-4 are exquisitely co-localized with phosphorylated Ephrin B at the point of septation in the tracheoesophageal foregut of developing mouse embryo.Citation70
On the opposite side of the Eph receptor-Ephrin pair, claudin-4 is also known to interact with the extracellular sequences EphA2.Citation71 Interaction leads to phosphorylation of the cytoplasmic tail of claudin-4 at Tyr-208 within the PDZ binding motif and this correlates loss of claudin-4 from the junction and increased barrier leakiness. More work is required but this and similar cell culture studies suggest a mechanism by which tumors that overexpress Eph receptors result in junction breakdown which precedes metastasis.
Unanswered Questions
It is clear that claudins have multiple interactions with many proteins, but a number of critical unanswered questions remain obstacles in our understanding of claudin associations. These include: (1) What is the three dimensional structure of a claudin? To date claudin crystallization attempts for X-ray diffraction or cryo-EM studies have not been successful. However, a recent report of a cryo-EM structure of a claudin related protein from Euglena gracilis, IP39Citation72 may provide insight into claudin organization in the membrane. This protein forms trimeric units which polymerized into strands. The trimer was asymmetric and could interact with neighbors in multiple ways. The authors postulated that combinations of similar multiple different interactions might allow strand formation. This structure may provide a template for understanding claudin structure and inform mutagenesis studies of claudins. (2) What interactions create the 10nM particle? Freeze fracture electron microscopy of unfixed tight junctions reveals not continuous tight junction strands, but rows of 10nm particles.Citation3 A critical remaining question is what are the components of these particles (presumably claudins, but potentially other proteins as well) and how are they organized to form this fundamental particle unit and continuous barrier forming rows? (3) How do claudins interact in cis and trans to create charge-selective pores? Beyond our own early studies,Citation11,Citation73 Yu and coworkersCitation74-Citation77 in an elegant series of studies have begun to define critical residues in claudin-2 that influence paracellular selectivity. Combined with structural studies, this type of analysis will help explain the organization of the pores. (4) How are claudin interactions regulated by post-translational modifications? Claudins are phosphorylated and palmitoylated (reviewed in ref. Citation78) but how this might affect claudin interactions or regulation is incompletely understood. (5) Finally, it is highly probable that claudins interact with many other proteins than those described above, including of course signaling and trafficking proteins as well as tight junction and lateral membrane proteins. The rules regulating the associations of these proteins are likely to provide insights into the whole life cycle of claudin and tight junction regulation and behavior.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work is supported by the Intramural Program of the National Heart, Lung and Blood Institute, National Institutes of Health, Bethesda, MD.
References
- Farquhar MG, Palade GE. Junctional complexes in various epithelia. J Cell Biol 1963; 17:375 - 412; http://dx.doi.org/10.1083/jcb.17.2.375; PMID: 13944428
- Chalcroft JP, Bullivant S. An interpretation of liver cell membrane and junction structure based on observation of freeze-fracture replicas of both sides of the fracture. J Cell Biol 1970; 47:49 - 60; http://dx.doi.org/10.1083/jcb.47.1.49; PMID: 4935338
- Staehelin LA. Further observations on the fine structure of freeze-cleaved tight junctions. J Cell Sci 1973; 13:763 - 86; PMID: 4203962
- Furuse M, Sasaki H, Fujimoto K, Tsukita S. A single gene product, claudin-1 or -2, reconstitutes tight junction strands and recruits occludin in fibroblasts. J Cell Biol 1998; 143:391 - 401; http://dx.doi.org/10.1083/jcb.143.2.391; PMID: 9786950
- Mineta K, Yamamoto Y, Yamazaki Y, Tanaka H, Tada Y, Saito K, et al. Predicted expansion of the claudin multigene family. FEBS Lett 2011; 585:606 - 12; http://dx.doi.org/10.1016/j.febslet.2011.01.028; PMID: 21276448
- Günzel D, Yu AS. Claudins and the modulation of tight junction permeability. Physiol Rev 2013; 93:525 - 69; http://dx.doi.org/10.1152/physrev.00019.2012; PMID: 23589827
- Nunes FD, Lopez LN, Lin HW, Davies C, Azevedo RB, Gow A, et al. Distinct subdomain organization and molecular composition of a tight junction with adherens junction features. J Cell Sci 2006; 119:4819 - 27; http://dx.doi.org/10.1242/jcs.03233; PMID: 17130295
- Ebrahim S, Fujita T, Millis BA, Kozin E, Ma X, Kawamoto S, et al. NMII Forms a Contractile Transcellular Sarcomeric Network to Regulate Apical Cell Junctions and Tissue Geometry. Curr Biol 2013; 23:731 - 6; http://dx.doi.org/10.1016/j.cub.2013.03.039; PMID: 23562268
- Van Itallie C, Rahner C, Anderson JM. Regulated expression of claudin-4 decreases paracellular conductance through a selective decrease in sodium permeability. J Clin Invest 2001; 107:1319 - 27; http://dx.doi.org/10.1172/JCI12464; PMID: 11375422
- Van Itallie CM, Gambling TM, Carson JL, Anderson JM. Palmitoylation of claudins is required for efficient tight-junction localization. J Cell Sci 2005; 118:1427 - 36; http://dx.doi.org/10.1242/jcs.01735; PMID: 15769849
- Colegio OR, Van Itallie C, Rahner C, Anderson JM. Claudin extracellular domains determine paracellular charge selectivity and resistance but not tight junction fibril architecture. Am J Physiol Cell Physiol 2003; 284:C1346 - 54; http://dx.doi.org/10.1152/ajpcell.00547.2002; PMID: 12700140
- McCarthy KM, Francis SA, McCormack JM, Lai J, Rogers RA, Skare IB, et al. Inducible expression of claudin-1-myc but not occludin-VSV-G results in aberrant tight junction strand formation in MDCK cells. J Cell Sci 2000; 113:3387 - 98; PMID: 10984430
- Sengoku A, Inai T, Shibata Y. Formation of aberrant TJ strands by overexpression of claudin-15 in MDCK II cells. Histochem Cell Biol 2008; 129:211 - 22; http://dx.doi.org/10.1007/s00418-007-0354-y; PMID: 17989991
- Fontijn RD, Rohlena J, van Marle J, Pannekoek H, Horrevoets AJ. Limited contribution of claudin-5-dependent tight junction strands to endothelial barrier function. Eur J Cell Biol 2006; 85:1131 - 44; http://dx.doi.org/10.1016/j.ejcb.2006.07.005; PMID: 16959372
- Furuse M, Sasaki H, Tsukita S. Manner of interaction of heterogeneous claudin species within and between tight junction strands. J Cell Biol 1999; 147:891 - 903; http://dx.doi.org/10.1083/jcb.147.4.891; PMID: 10562289
- Mitic LL, Unger VM, Anderson JM. Expression, solubilization, and biochemical characterization of the tight junction transmembrane protein claudin-4. Protein Sci 2003; 12:218 - 27; http://dx.doi.org/10.1110/ps.0233903; PMID: 12538885
- Van Itallie CM, Mitic LL, Anderson JM. Claudin-2 forms homodimers and is a component of a high molecular weight protein complex. J Biol Chem 2011; 286:3442 - 50; http://dx.doi.org/10.1074/jbc.M110.195578; PMID: 21098027
- Yamazaki Y, Tokumasu R, Kimura H, Tsukita S. Role of claudin species-specific dynamics in reconstitution and remodeling of the zonula occludens. Mol Biol Cell 2011; 22:1495 - 504; http://dx.doi.org/10.1091/mbc.E10-12-1003; PMID: 21372174
- Inai T, Kamimura T, Hirose E, Iida H, Shibata Y. The protoplasmic or exoplasmic face association of tight junction particles cannot predict paracellular permeability or heterotypic claudin compatibility. Eur J Cell Biol 2010; 89:547 - 56; http://dx.doi.org/10.1016/j.ejcb.2010.01.003; PMID: 20188437
- Inai T, Sengoku A, Hirose E, Iida H, Shibata Y. Claudin-7 expressed on lateral membrane of rat epididymal epithelium does not form aberrant tight junction strands. Anat Rec (Hoboken) 2007; 290:1431 - 8; http://dx.doi.org/10.1002/ar.20597; PMID: 17853415
- Ikenouchi J, Sasaki H, Tsukita S, Furuse M, Tsukita S. Loss of occludin affects tricellular localization of tricellulin. Mol Biol Cell 2008; 19:4687 - 93; http://dx.doi.org/10.1091/mbc.E08-05-0530; PMID: 18768749
- Piontek J, Fritzsche S, Cording J, Richter S, Hartwig J, Walter M, et al. Elucidating the principles of the molecular organization of heteropolymeric tight junction strands. Cell Mol Life Sci 2011; 68:3903 - 18; http://dx.doi.org/10.1007/s00018-011-0680-z; PMID: 21533891
- Sekar RB, Periasamy A. Fluorescence resonance energy transfer (FRET) microscopy imaging of live cell protein localizations. J Cell Biol 2003; 160:629 - 33; http://dx.doi.org/10.1083/jcb.200210140; PMID: 12615908
- Hou J, Renigunta A, Gomes AS, Hou M, Paul DL, Waldegger S, et al. Claudin-16 and claudin-19 interaction is required for their assembly into tight junctions and for renal reabsorption of magnesium. Proc Natl Acad Sci U S A 2009; 106:15350 - 5; http://dx.doi.org/10.1073/pnas.0907724106; PMID: 19706394
- Hou J, Renigunta A, Yang J, Waldegger S. Claudin-4 forms paracellular chloride channel in the kidney and requires claudin-8 for tight junction localization. Proc Natl Acad Sci U S A 2010; 107:18010 - 5; http://dx.doi.org/10.1073/pnas.1009399107; PMID: 20921420
- Piehl C, Piontek J, Cording J, Wolburg H, Blasig IE. Participation of the second extracellular loop of claudin-5 in paracellular tightening against ions, small and large molecules. Cell Mol Life Sci 2010; 67:2131 - 40; http://dx.doi.org/10.1007/s00018-010-0332-8; PMID: 20333434
- Piontek J, Winkler L, Wolburg H, Müller SL, Zuleger N, Piehl C, et al. Formation of tight junction: determinants of homophilic interaction between classic claudins. FASEB J 2008; 22:146 - 58; http://dx.doi.org/10.1096/fj.07-8319com; PMID: 17761522
- Lim TS, Vedula SR, Hunziker W, Lim CT. Kinetics of adhesion mediated by extracellular loops of claudin-2 as revealed by single-molecule force spectroscopy. J Mol Biol 2008; 381:681 - 91; http://dx.doi.org/10.1016/j.jmb.2008.06.009; PMID: 18635194
- Daugherty BL, Ward C, Smith T, Ritzenthaler JD, Koval M. Regulation of heterotypic claudin compatibility. J Biol Chem 2007; 282:30005 - 13; http://dx.doi.org/10.1074/jbc.M703547200; PMID: 17699514
- Kubota K, Furuse M, Sasaki H, Sonoda N, Fujita K, Nagafuchi A, et al. Ca(2+)-independent cell-adhesion activity of claudins, a family of integral membrane proteins localized at tight junctions. Curr Biol 1999; 9:1035 - 8; http://dx.doi.org/10.1016/S0960-9822(99)80452-7; PMID: 10508613
- Matsuda M, Kubo A, Furuse M, Tsukita S. A peculiar internalization of claudins, tight junction-specific adhesion molecules, during the intercellular movement of epithelial cells. J Cell Sci 2004; 117:1247 - 57; http://dx.doi.org/10.1242/jcs.00972; PMID: 14996944
- Shen L, Weber CR, Turner JR. The tight junction protein complex undergoes rapid and continuous molecular remodeling at steady state. J Cell Biol 2008; 181:683 - 95; http://dx.doi.org/10.1083/jcb.200711165; PMID: 18474622
- Sasaki H, Matsui C, Furuse K, Mimori-Kiyosue Y, Furuse M, Tsukita S. Dynamic behavior of paired claudin strands within apposing plasma membranes. Proc Natl Acad Sci U S A 2003; 100:3971 - 6; http://dx.doi.org/10.1073/pnas.0630649100; PMID: 12651952
- Furuse M, Hirase T, Itoh M, Nagafuchi A, Yonemura S, Tsukita S, et al. Occludin: a novel integral membrane protein localizing at tight junctions. J Cell Biol 1993; 123:1777 - 88; http://dx.doi.org/10.1083/jcb.123.6.1777; PMID: 8276896
- Saitou M, Fujimoto K, Doi Y, Itoh M, Fujimoto T, Furuse M, et al. Occludin-deficient embryonic stem cells can differentiate into polarized epithelial cells bearing tight junctions. J Cell Biol 1998; 141:397 - 408; http://dx.doi.org/10.1083/jcb.141.2.397; PMID: 9548718
- Cording J, Berg J, Käding N, Bellmann C, Tscheik C, Westphal JK, et al. In tight junctions, claudins regulate the interactions between occludin, tricellulin and marvelD3, which, inversely, modulate claudin oligomerization. J Cell Sci 2013; 126:554 - 64; http://dx.doi.org/10.1242/jcs.114306; PMID: 23203797
- Ikenouchi J, Furuse M, Furuse K, Sasaki H, Tsukita S, Tsukita S. Tricellulin constitutes a novel barrier at tricellular contacts of epithelial cells. J Cell Biol 2005; 171:939 - 45; http://dx.doi.org/10.1083/jcb.200510043; PMID: 16365161
- Raleigh DR, Marchiando AM, Zhang Y, Shen L, Sasaki H, Wang Y, et al. Tight junction-associated MARVEL proteins marveld3, tricellulin, and occludin have distinct but overlapping functions. Mol Biol Cell 2010; 21:1200 - 13; http://dx.doi.org/10.1091/mbc.E09-08-0734; PMID: 20164257
- Raleigh DR, Boe DM, Yu D, Weber CR, Marchiando AM, Bradford EM, et al. Occludin S408 phosphorylation regulates tight junction protein interactions and barrier function. J Cell Biol 2011; 193:565 - 82; http://dx.doi.org/10.1083/jcb.201010065; PMID: 21536752
- Harris HJ, Davis C, Mullins JG, Hu K, Goodall M, Farquhar MJ, et al. Claudin association with CD81 defines hepatitis C virus entry. J Biol Chem 2010; 285:21092 - 102; http://dx.doi.org/10.1074/jbc.M110.104836; PMID: 20375010
- Mrsny RJ, Brown GT, Gerner-Smidt K, Buret AG, Meddings JB, Quan C, et al. A key claudin extracellular loop domain is critical for epithelial barrier integrity. Am J Pathol 2008; 172:905 - 15; http://dx.doi.org/10.2353/ajpath.2008.070698; PMID: 18349130
- Yu AS, McCarthy KM, Francis SA, McCormack JM, Lai J, Rogers RA, et al. Knockdown of occludin expression leads to diverse phenotypic alterations in epithelial cells. Am J Physiol Cell Physiol 2005; 288:C1231 - 41; http://dx.doi.org/10.1152/ajpcell.00581.2004; PMID: 15689410
- Van Itallie CM, Fanning AS, Holmes J, Anderson JM. Occludin is required for cytokine-induced regulation of tight junction barriers. J Cell Sci 2010; 123:2844 - 52; http://dx.doi.org/10.1242/jcs.065581; PMID: 20663912
- Beeman N, Webb PG, Baumgartner HK. Occludin is required for apoptosis when claudin-claudin interactions are disrupted. Cell Death Dis 2012; 3:e273; http://dx.doi.org/10.1038/cddis.2012.14; PMID: 22361748
- Fanning AS, Anderson JM. Zonula occludens-1 and -2 are cytosolic scaffolds that regulate the assembly of cellular junctions. Ann N Y Acad Sci 2009; 1165:113 - 20; http://dx.doi.org/10.1111/j.1749-6632.2009.04440.x; PMID: 19538295
- Morita K, Furuse M, Fujimoto K, Tsukita S. Claudin multigene family encoding four-transmembrane domain protein components of tight junction strands. Proc Natl Acad Sci U S A 1999; 96:511 - 6; http://dx.doi.org/10.1073/pnas.96.2.511; PMID: 9892664
- Itoh M, Furuse M, Morita K, Kubota K, Saitou M, Tsukita S. Direct binding of three tight junction-associated MAGUKs, ZO-1, ZO-2, and ZO-3, with the COOH termini of claudins. J Cell Biol 1999; 147:1351 - 63; http://dx.doi.org/10.1083/jcb.147.6.1351; PMID: 10601346
- Fanning AS, Van Itallie CM, Anderson JM. Zonula occludens-1 and -2 regulate apical cell structure and the zonula adherens cytoskeleton in polarized epithelia. Mol Biol Cell 2012; 23:577 - 90; http://dx.doi.org/10.1091/mbc.E11-09-0791; PMID: 22190737
- Ikenouchi J, Umeda K, Tsukita S, Furuse M, Tsukita S. Requirement of ZO-1 for the formation of belt-like adherens junctions during epithelial cell polarization. J Cell Biol 2007; 176:779 - 86; http://dx.doi.org/10.1083/jcb.200612080; PMID: 17353356
- Rüffer C, Gerke V. The C-terminal cytoplasmic tail of claudins 1 and 5 but not its PDZ-binding motif is required for apical localization at epithelial and endothelial tight junctions. Eur J Cell Biol 2004; 83:135 - 44; http://dx.doi.org/10.1078/0171-9335-00366; PMID: 15260435
- Van Itallie CM, Colegio OR, Anderson JM. The cytoplasmic tails of claudins can influence tight junction barrier properties through effects on protein stability. J Membr Biol 2004; 199:29 - 38; http://dx.doi.org/10.1007/s00232-004-0673-z; PMID: 15366421
- Müller D, Kausalya PJ, Claverie-Martin F, Meij IC, Eggert P, Garcia-Nieto V, et al. A novel claudin 16 mutation associated with childhood hypercalciuria abolishes binding to ZO-1 and results in lysosomal mistargeting. Am J Hum Genet 2003; 73:1293 - 301; http://dx.doi.org/10.1086/380418; PMID: 14628289
- Appleton BA, Zhang Y, Wu P, Yin JP, Hunziker W, Skelton NJ, et al. Comparative structural analysis of the Erbin PDZ domain and the first PDZ domain of ZO-1. Insights into determinants of PDZ domain specificity. J Biol Chem 2006; 281:22312 - 20; http://dx.doi.org/10.1074/jbc.M602901200; PMID: 16737969
- Jeansonne B, Lu Q, Goodenough DA, Chen YH. Claudin-8 interacts with multi-PDZ domain protein 1 (MUPP1) and reduces paracellular conductance in epithelial cells. Cell Mol Biol (Noisy-le-grand) 2003; 49:13 - 21; PMID: 12839333
- Lanaspa MA, Andres-Hernando A, Rivard CJ, Dai Y, Berl T. Hypertonic stress increases claudin-4 expression and tight junction integrity in association with MUPP1 in IMCD3 cells. Proc Natl Acad Sci U S A 2008; 105:15797 - 802; http://dx.doi.org/10.1073/pnas.0805761105; PMID: 18840681
- Poliak S, Matlis S, Ullmer C, Scherer SS, Peles E. Distinct claudins and associated PDZ proteins form different autotypic tight junctions in myelinating Schwann cells. J Cell Biol 2002; 159:361 - 72; http://dx.doi.org/10.1083/jcb.200207050; PMID: 12403818
- Roh MH, Liu CJ, Laurinec S, Margolis B. The carboxyl terminus of zona occludens-3 binds and recruits a mammalian homologue of discs lost to tight junctions. J Biol Chem 2002; 277:27501 - 9; http://dx.doi.org/10.1074/jbc.M201177200; PMID: 12021270
- Munz M, Baeuerle PA, Gires O. The emerging role of EpCAM in cancer and stem cell signaling. Cancer Res 2009; 69:5627 - 9; http://dx.doi.org/10.1158/0008-5472.CAN-09-0654; PMID: 19584271
- Ladwein M, Pape UF, Schmidt DS, Schnölzer M, Fiedler S, Langbein L, et al. The cell-cell adhesion molecule EpCAM interacts directly with the tight junction protein claudin-7. Exp Cell Res 2005; 309:345 - 57; http://dx.doi.org/10.1016/j.yexcr.2005.06.013; PMID: 16054130
- Kuhn S, Koch M, Nübel T, Ladwein M, Antolovic D, Klingbeil P, et al. A complex of EpCAM, claudin-7, CD44 variant isoforms, and tetraspanins promotes colorectal cancer progression. Mol Cancer Res 2007; 5:553 - 67; http://dx.doi.org/10.1158/1541-7786.MCR-06-0384; PMID: 17579117
- Nübel T, Preobraschenski J, Tuncay H, Weiss T, Kuhn S, Ladwein M, et al. Claudin-7 regulates EpCAM-mediated functions in tumor progression. Mol Cancer Res 2009; 7:285 - 99; http://dx.doi.org/10.1158/1541-7786.MCR-08-0200; PMID: 19276185
- Lei Z, Maeda T, Tamura A, Nakamura T, Yamazaki Y, Shiratori H, et al. EpCAM contributes to formation of functional tight junction in the intestinal epithelium by recruiting claudin proteins. Dev Biol 2012; 371:136 - 45; http://dx.doi.org/10.1016/j.ydbio.2012.07.005; PMID: 22819673
- Wu CJ, Mannan P, Lu M, Udey MC. Epithelial Cell Adhesion Molecule (EpCAM) Regulates Claudin Dynamics and Tight Junctions. J Biol Chem 2013; 288:12253 - 68; http://dx.doi.org/10.1074/jbc.M113.457499; PMID: 23486470
- Rubinstein E. The complexity of tetraspanins. Biochem Soc Trans 2011; 39:501 - 5; http://dx.doi.org/10.1042/BST0390501; PMID: 21428928
- Tiwari-Woodruff SK, Buznikov AG, Vu TQ, Micevych PE, Chen K, Kornblum HI, et al. OSP/claudin-11 forms a complex with a novel member of the tetraspanin super family and beta1 integrin and regulates proliferation and migration of oligodendrocytes. J Cell Biol 2001; 153:295 - 305; http://dx.doi.org/10.1083/jcb.153.2.295; PMID: 11309411
- Kovalenko OV, Yang XH, Hemler ME. A novel cysteine cross-linking method reveals a direct association between claudin-1 and tetraspanin CD9. Mol Cell Proteomics 2007; 6:1855 - 67; http://dx.doi.org/10.1074/mcp.M700183-MCP200; PMID: 17644758
- Stamataki Z, Grove J, Balfe P, McKeating JA. Hepatitis C virus entry and neutralization. [x.] Clin Liver Dis 2008; 12:693 - 712, x; http://dx.doi.org/10.1016/j.cld.2008.03.008; PMID: 18625435
- Himanen JP, Saha N, Nikolov DB. Cell-cell signaling via Eph receptors and ephrins. Curr Opin Cell Biol 2007; 19:534 - 42; http://dx.doi.org/10.1016/j.ceb.2007.08.004; PMID: 17928214
- Tanaka M, Kamata R, Sakai R. Phosphorylation of ephrin-B1 via the interaction with claudin following cell-cell contact formation. EMBO J 2005; 24:3700 - 11; http://dx.doi.org/10.1038/sj.emboj.7600831; PMID: 16211011
- Dravis C, Henkemeyer M. Ephrin-B reverse signaling controls septation events at the embryonic midline through separate tyrosine phosphorylation-independent signaling avenues. Dev Biol 2011; 355:138 - 51; http://dx.doi.org/10.1016/j.ydbio.2011.04.020; PMID: 21539827
- Tanaka M, Kamata R, Sakai R. EphA2 phosphorylates the cytoplasmic tail of Claudin-4 and mediates paracellular permeability. J Biol Chem 2005; 280:42375 - 82; http://dx.doi.org/10.1074/jbc.M503786200; PMID: 16236711
- Suzuki H, Ito Y, Yamazaki Y, Mineta K, Uji M, Abe K, et al. The four-transmembrane protein IP39 of Euglena forms strands by a trimeric unit repeat. Nat Commun 2013; 4:1766; http://dx.doi.org/10.1038/ncomms2731; PMID: 23612307
- Colegio OR, Van Itallie CM, McCrea HJ, Rahner C, Anderson JM. Claudins create charge-selective channels in the paracellular pathway between epithelial cells. Am J Physiol Cell Physiol 2002; 283:C142 - 7; http://dx.doi.org/10.1152/ajpcell.00038.2002; PMID: 12055082
- Angelow S, Yu AS. Cysteine mutagenesis to study the structure of claudin-2 paracellular pores. Ann N Y Acad Sci 2009; 1165:143 - 7; http://dx.doi.org/10.1111/j.1749-6632.2009.04038.x; PMID: 19538299
- Angelow S, Yu AS. Structure-function studies of claudin extracellular domains by cysteine-scanning mutagenesis. J Biol Chem 2009; 284:29205 - 17; http://dx.doi.org/10.1074/jbc.M109.043752; PMID: 19690347
- Yu AS, Cheng MH, Angelow S, Günzel D, Kanzawa SA, Schneeberger EE, et al. Molecular basis for cation selectivity in claudin-2-based paracellular pores: identification of an electrostatic interaction site. J Gen Physiol 2009; 133:111 - 27; http://dx.doi.org/10.1085/jgp.200810154; PMID: 19114638
- Yu AS, Cheng MH, Coalson RD. Calcium inhibits paracellular sodium conductance through claudin-2 by competitive binding. J Biol Chem 2010; 285:37060 - 9; http://dx.doi.org/10.1074/jbc.M110.146621; PMID: 20807759
- Findley MK, Koval M. Regulation and roles for claudin-family tight junction proteins. IUBMB Life 2009; 61:431 - 7; http://dx.doi.org/10.1002/iub.175; PMID: 19319969