Abstract
The recognition of T cell intracellular antigen-1 (TIA-1) by Fas-activated Ser/Thr phosphoprotein (FAST) results in prolonged cell survival by inducing the expression of inhibitors of apoptosis. Here we show that the functional effects of FAST are dependent on its interactions with eukaryotic translation initiation factor 4E (eIF4E) which is the major cytosolic cap binding protein in cells. FAST binds to eIF4E via a consensus motif (428YXXXXLL433) that is also found in eIF4G, 4E-BP1/2/3, 4E-T, and cup. A point mutation within this motif at Y428 dampens the ability of FAST to recognize eIF4E. Wild-type (WT) FAST, but not its Y428G mutant, increases the expression of co-transfected cellular inhibitor of apoptosis-1 (cIAP-1) and β-gal mRNA and protein, but inhibits the Fas-induced activation of caspase-3. Increased expression of the co-transfected proteins results, in part, from stabilization of mRNA, suggesting that FAST:eIF4E interactions can inhibit mRNA decay. We propose that eIF4E:FAST:TIA-1 complexes regulate the translation and stability of specific mRNAs that encode proteins important for cell survival.
Keywords: :
Introduction
The RNA-binding proteins T cell intracellular antigen-1 (TIA-1) and its related protein TIAR/TIAL1 are translational silencers that preferentially target mRNAs bearing adenine/uridine-rich elements (AREs) such as the AUUUA motif in their 3′ untranslated regions.Citation1-Citation3 In cells subjected to environmental stress (e.g., heat, UV irradiation, and oxidative conditions), TIA-1 and TIAR contribute to a general translational arrest that is initiated by the phosphorylation of the translation initiation factor eIF2α. In response to phospho-eIF2α, TIA-1 and TIAR promote the assembly of a noncanonical 48S pre-initiation complex that inhibits protein translation.Citation4 These complexes accumulate at discrete cytoplasmic foci known as stress granules (SGs).Citation1 Because mRNA is shuttled in and out of SGs, these structures have been proposed to be sites at which mRNAs are monitored for composition, and then directed to sites of degradation, re-initiation, or storage.Citation1 SGs exhibit regulated interactions with processing bodies (PBs) which are distinct cytoplasmic structures that contain components of a 5′→3′ mRNA degradative machinery.Citation5,Citation6 The accumulated data suggest that mRNAs destined for degradation move from SGs to PBs.Citation7
Fas-activated Ser/Thr phosphoprotein (FAST) binds to TIA-1.Citation8 FAST is rapidly dephosphorylated in Jurkat cells that are undergoing Fas-induced apoptosis, suggesting an undefined role for the protein in apoptosis.Citation8 Consistent with this hypothesis, overexpression of FAST in cells results in reduced Fas-induced apoptosis.Citation9 Because the anti-apoptotic effects of FAST are inhibited by TIA-1, it has been concluded that FAST:TIA-1 interactions can regulate apoptotic cell death.Citation9 FAST promotes the expression of β-gal in co-transfected cells, as well as endogenous c-IAP-1 and XIAP.Citation9 Thus, the anti-apoptotic effects of FAST may be due to enhanced expression of one or more inhibitors of apoptosis. These results suggest that FAST and TIA-1 are functional antagonists that regulate protein expression and cell survival.
Like TIA-1, Drosophila melanogaster-derived cytoplasmic polyadenylation element binding protein (CPEB) and Bruno are translational silencers that bind to cis elements in the 3′ untranslated regions of their target transcripts.Citation10 These proteins inhibit translation by recruiting eukaryotic translation initiation factor 4E (eIF4E)-binding proteins (maskin and cup, respectively) that prevent the recruitment of eIF4G and the assembly of 48S pre-initiation complexes.Citation10 These translationally silenced mRNAs are concomitantly stabilized, making them available for subsequent re-initiation when conditions are favorable. Here we show that FAST is an eIF4E-binding protein that possesses two Y-X-X-X-X-L-F (where X is any residue and ϕ is Leu, Met or Phe) sequencesCitation11 that allow eIF4G, 4E-BP1, 4E-BP2, 4E-BP3, 4E-T, and cup to bind to eIF4E.Citation12-Citation16 These proteins inhibit translation by preventing the recruitment of eIF4G to the 48S pre-initiation complex.Citation17 We show that FAST similarly inhibits eIF4E:eIF4G interactions, suggesting that it may be a functional ortholog of maskin and cup.
In addition to regulating translational initiation, interactions between eIF4E and the 7-methyl guanine cap can regulate mRNA degradation. In the 5′→3′ mRNA decay pathway, the decapping enzymes DCP1 and DCP2 remove the 7-methyl guanine cap, allowing the 5′→3′ exonuclease Xrn1 to degrade the mRNA.Citation18 eIF4E inhibits this degradative pathway by preventing Dcp1/Dcp2-mediated decapping.Citation17 As a consequence, eIF4G and 4E-BPs can inhibit mRNA degradation by stabilizing the interactions between eIF4E and cap.Citation17 Here we show that FAST binds to eIF4E and inhibits mRNA degradation. In contrast, a Y428G mutant of FAST that no longer binds to eIF4E fails to prevent mRNA degradation. Moreover, this mutated isoform of FAST potentiates Fas-induced apoptosis, consistent with a role for FAST in regulating the expression of apoptotic regulatory proteins. Finally, our data show that FAST can bind to both eIF4E and TIA-1, suggesting that interactions between these translational control proteins may regulate mRNA stability, mRNA translation and cell survival.
Results
FAST recognizes eIF4E−Sequence analysis revealed that FAST possesses two potential eIF4E-binding consensus motifs. The sequence alignment of these possible eIF4E-binding sites is compared with those found in other eIF4E-binding partners in . To determine whether FAST can bind to eIF4E, we performed a co-immunoprecipitation analysis using recombinant HA-FAST and recombinant FLAG-eIF4E. COS-7 cells were co-transfected with pcDNA3-FLAG-eIF4E together with either vector control, pMT2-HA-WT-FAST, or pMT2-HA-Y428G-FAST (a point mutant in which the Tyr in the second eIF4E-binding motif is replaced with a Gly). After 28 h, cells were harvested for immunoprecipitation analysis using anti-HA Ab followed by immunoblotting with either anti-HA or anti-FLAG Ab. The results revealed that recombinant HA-FAST can efficiently co-precipitate recombinant FLAG-eIF4E (). Although HA-Y428G-FAST was efficiently precipitated using anti-HA, no FLAG-eIF4E was co-precipitated. These results revealed that recombinant FAST can bind to recombinant eIF4E and that Y428 is required for this interaction.
Figure 1. FAST encodes an eIF4E-binding motif. Amino acid sequence alignment comparing the eIF4E-binding motifs of FAST with those found in human eIF4GI (AF012088), human eIF4GII (AF012072), human 4E-BP1 (L36055), human 4E-BP2 (L36056), human 4E-BP3 (AF038869), human 4E-T (AF240775), Drosophila eIF4G (AF030155), S. cerevisiae eIF4GI (p39935), S. cerevisiae eIF4GII (p39936), S.cerevisiae p20 (X15731) and wheat iso-eIF4G (M95747). Residues which are critical for eIF4E binding are boxed and shaded. The additional identical or reserved residues are boxed.
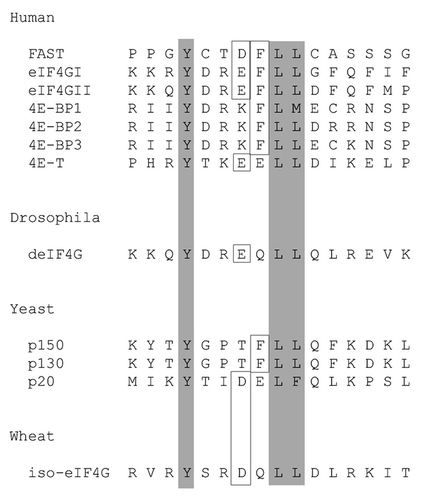
Figure 2.FAST interacts with eIF4E through its eIF4E-binding motif.A, FLAG-tagged eIF4E was overexpressed together with the indicated constructs in COS-7 cells. Cell lysates were immunoprecipitated with anti-HA Ab, then subjected to SDS-PAGE immunoblot analysis. Left upper panel: immunoblotting with anti-FLAG Ab after IP; Left lower panel: immunoblotting with anti-HA Ab after IP; Right upper panel: immunoblotting with anti-FLAG Ab before IP; Right lower panel: immunoblotting with anti-HA Ab before IP. B, HA-tagged WT-FAST and Y428G-FAST were overexpressed in HeLa cells. Cell lysates were immunoprecipitated with a mouse anti-eIF4E Ab followed by SDS-PAGE immunoblot analysis. Left panel: immunoblotting with anti-HA Ab after immunopreciptitation; Right panel: immunoblotting with anti-HA Ab before immunoprecipitation.
To determine whether recombinant FAST can bind to endogenous eIF4E, HeLa cells were transfected with either pMT2-HA-WT FAST or pMT2-HA-Y428G-FAST, and then were processed for immunoprecipitation analysis using an anti-eIF4E Ab. Although HA-WT-FAST and HA-Y428G-FAST were expressed at similar levels, eIF4E co-precipitated HA-WT- FAST, but not its Y428G mutant (). Together, these data demonstrate that endogenous eIF4E can bind to recombinant FAST via its minimal eIF4E-binding motif.
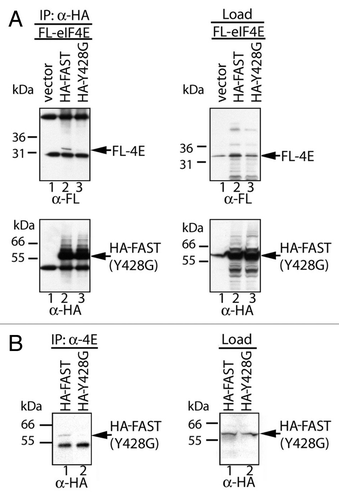
The ability of HA-FAST to bind endogenous eIF4E was confirmed in U2OS cells transfected with HA-FAST (WT) or HA-Y428G-FAST (MUT). Transfected cells were lysed in the absence or presence of RNase A and immunoprecipitated with mouse IgG antibodies or anti-eIF4E antibodies before separating the load and immunoprecitates by SDS-PAGE. Subsequent immunoblotting analysis was used to quantify endogenous eIF4E and transfected HA-tagged recombinant proteins. As shown in , HA-FAST, but not HA-Y428G-FAST is co-precipitated with eIF4E in the absence or presence of RNase. This result indicates that RNA is not required for interations between eIF4E and HA-FAST.
Figure 3. FAST:eIF4E interactions are resistant to RNase. U2OS cells were transfected with pMT2-HA vector (-) or pMT2-HA-FAST encoding wild type (WT) or Y428G (MUT) variants. Transfected cells were lysed in the presence or absence of RNase A as indicated, and subjected to immunoprecipitation using mouse IgG antibodies (IP:IgG) or eIF4E (IP:eIF4E) antibodies. Co-immunoprecipitation efficiency was quantified by immunoblotting using anti-HA and anti-eIF4E antibodies. Load: loading control (1/20th of total protein lysate).
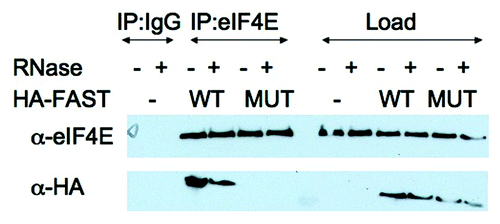
The specificity of eIF4E:HA-FAST interactions was further confirmed in a m7GTP pull down analysis. U2OS cells transfected with HA-FAST (WT) or HA-Y428G-FAST (MUT) were lysed in the presence of RNase and subjected to pull-down analysis using Sepharose-m7GTP beads. Proteins were quantified by immunoblotting analysis using anti-HA and anti-eIF4E. As shown in , m7GTP beads pull down similar amounts of endogenous eIF4E under all conditions. In contrast, WT but not mutant HA-FAST is pulled down by these beads. This result supports our contention that the Y428G point mutation eliminates binding to eIF4E.
Figure 4. m7GTP cap pull-down analysis. U2OS cells were transfected with pMT2-HA vector (-) or pMT2-HA-FAST encoding wild type (WT) or Y428G (MUT) variants. Transfected cells were lysed in the presence of RNase A and subjected to pull-down using m7GTP Sepharose (m7GTP). Pulled-down proteins were quantified by immunoblotting using anti-HA and anti-eIF4E antibodies. Load: loading control (1/20th of total protein lysate)
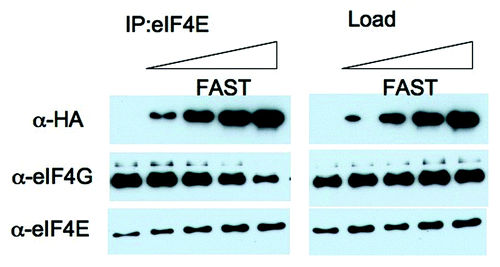
FAST displaces eIF4G from eIF4E−The known eIF4E-binding proteins inhibit translation by displacing eIF4G from eIF4E. To determine whether FAST similarly displaces eIF4G from eIF4E, we transfected U2OS cells with increasing amounts of HA-FAST before immunoprecipitating eIF4E from cell lysates. Cell lysates and immunoprecipitates were separated by SDS-PAGE, transferred to nitrocellulose membranes and blotted with anti-HA, anti-eIF4G, and anti-eIF4E (). In the presence of increasing amounts of HA-FAST, the amount of eIF4G that co-precipitates with eIF4E is progressively reduced (, left panel). This result suggests that FAST, like 4E-BP, can displace eIF4G from eIF4E.
Figure 5.FAST displaces eIF4G from eIF4E. U2OS cells were transfected with pMT2-HA vector (lane 1) or with an increasing amount of pMT2-HA-FAST encoding wild type FAST protein. Transfected cells were lysed in the presence of RNase A, and subjected to immunoprecipitation using eIF4E antibodies (IP:eIF4E). Co-immunoprecipitation efficiency was quantified by immunoblotting using anti-HA, anti-eIF4G and anti-eIF4E antibodies. Load: loading control (1/20th of total protein lysate).
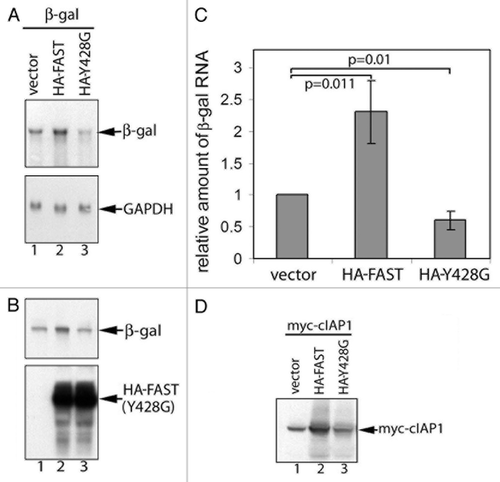
FAST regulates mRNA stability−We previously reported that recombinant FAST increases the expression of co-transfected β-gal.Citation9 Because FAST interacts with the translational silencer TIA-1, we speculated that this was a consequence of enhanced protein translation.Citation9 However, the ability of FAST to bind to eIF4E suggested that, like eIF4G and 4E-BP, it might also regulate mRNA stability. To test this hypothesis, COS-7 cells were co-transfected with the reporter plasmid pcDNA3-β-gal and either pMT2-HA-WT-FAST or pMT2-HA-Y428G-FAST. After 48 h, the transfected cells were harvested to obtain total RNA for RNA gel blot analysis or total protein for SDS-immunoblot analysis. The expression of β-gal mRNA ( and ) and protein () were consistently and significantly elevated in FAST transfectants. In contrast, the Y428G mutant significantly reduced the expression of β-gal mRNA ( and ). Unlike WT FAST, its Y428G mutant was not able to increase the expression of β-gal protein compared with vector control (). Similar results were obtained when Myc-cIAP-1 was used as a reporter protein in this assay (), consistent with previous findings.Citation9
Figure 6.FAST and FAST (Y428G) have different effects on the expression of co-transfected β-gal and cIAP-1. β-gal (A and B) or Myc-cIAP-1 (D) was expressed together with the indicated constructs in COS-7 cells. Total cellular RNA (A) or total proteins (B and D) were extracted and subjected to RNA gel blot and SDS-PAGE immunoblot analysis, respectively. (C) The mean relative amount of β-gal mRNA in each transfectant was quantified using densitometry and presented as a bar graph (± standard errors, n = 3). The amount of β-gal mRNA expressed in cells transfected with the vector control was arbitrarily designated as 1. The relative amount of β-gal mRNA expressed in cells transfected with HA-FAST or HA-FAST (Y428G) was calculated by dividing their absolute expression levels by the absolute expression level in cells transfected with the vector control. Calculated P values are shown for selected samples.
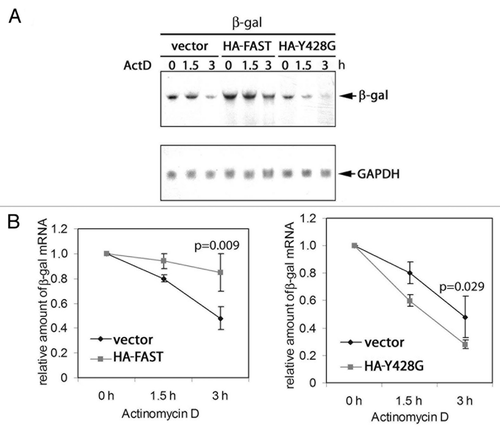
We next determined the half-life of β-gal mRNA in cells transfected with vector, HA-WT FAST, or HA-Y428G-FAST. COS cells were co-transfected with pcDNA3-β-gal together with either vector control, pMT2-HA-WT-FAST, or pMT2-HA-Y428G-FAST. After 48 h, cells were treated with actinomycin D to inhibit mRNA synthesis. The treated cells were harvested after 0, 1.5 or 3 h and processed for RNA gel blotting to quantify the expression of β-gal mRNA. A representative experiment is shown in . Decay curves calculated using data obtained from 3 independent experiments are shown in . The results revealed that FAST stabilizes, and Y428G-FAST destabilizes, β-gal mRNA. These results suggest that Y428G FAST somehow functions as a dominant negative inhibitor of endogenous FAST in these assays.
Figure 7.FAST and FAST (Y428G) have different effects on the stability of co-transfected β-gal transcripts.(A) COS-7 cells were co-transfected with β-gal together with either WT FAST or Y428G FAST. After 48 h, cells were treated with actinomycin D (5 mg/ml) for 0, 1.5, or 3 h. Total RNA was extracted and subjected to blot analysis.(B)The mean relative amounts of β-gal mRNA is plotted as a function of time in the presence of actinomycin D (± standard errors, n = 3). The relative amount of β-gal mRNA from samples treated with actinomycin D for 0 h (untreated) was arbitrarily designated as 1. The relative amount of the β-gal mRNA from samples treated with actinomycin D for 1.5 or 3 h was calculated by dividing their absolute values by the absolute amount of the β-gal mRNA of their corresponding untreated samples. In both cases, the t1/2 of β-gal mRNA in vector transfected cells (right panel) was 2.9. In contrast, the t1/2 of β-gal mRNA in HA-FAST transfected cells (left panel) was 9.4 and the t1/2 of β-gal mRNA in HA-Y428G transfected cells was 1.6. Calculated P values are shown for the samples treated with actinomycin D for 3 h.
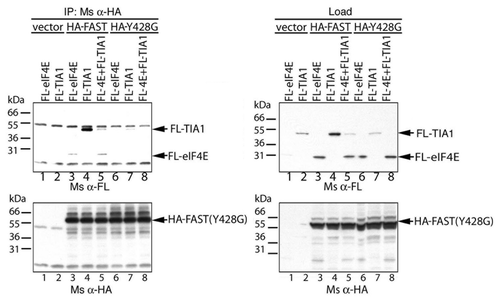
The Y428G mutant of FAST retains its ability to bind to TIA-1−We previously showed that TIA-1 binds to FAST (1–372), a truncation mutant that lacks the 428YCTDFLL434 eIF4E-binding motif. This was confirmed in the co-immunoprecipitation analysis shown in . COS-7 cells were co-transfected with either vector control, pMT2-HA-WT-FAST, or pMT2-HA-Y428G-FAST together with either pcDNA3-FLAG-eIF4E and/or pcDNA3-FLAG-TIA1, cultured for 28 h, then processed for co-immunoprecipitation analysis using anti-HA Ab. The results showed that both WT FAST and Y428G FAST co-precipitated FLAG-TIA1 (). Much more FLAG-TIA1 co-precipitated with WT FAST than its mutant (). However, the expression of FLAG-TIA1 was much less when co-expressed with Y428G FAST compared with WT FAST (). Our data therefore showed that Y428G-FAST retains the ability to bind to TIA-1. When HA-WT-FAST is co-expressed with FLAG-TIA1 and FLAG-eIF4E, both proteins co-precipitate with HA-WT-FAST (). This result suggests that the binding of TIA-1 does not prevent the binding of eIF4E.
Figure 8.The Y428G mutant of FAST binds TIA-1. FLAG-tagged eIF4E and/or TIA-1 was overexpressed together with the indicated constructs in COS-7 cells. Cell lysates were immunoprecipitated with anti-HA Ab, then subjected to SDS-PAGE immunoblot analysis. Upper left panel: immunoblotting with anti-FLAG Ab after IP; Upper right panel: immunoblotting with anti-FLAG Ab before immunoprecioutation; Lower left panel: immunoblotting with anti-HA Ab after i.p.; Lower right panel: immunoblotting with anti-HA Ab before immunoprecipitation
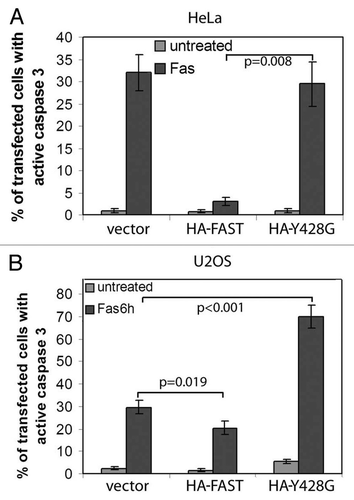
WT- and Y428G-FAST have different effects on Fas-induced apoptosis−We previously reported that the ability of FAST to increase the expression of a co-transfected reporter gene correlates with its ability to protect cells from Fas-induced apoptosis.Citation9 Since Y428G-FAST does not increase the expression of co-transfected β-gal or c-IAP-1, we predicted that it would not inhibit Fas-induced apoptosis. To test this prediction, HeLa and U2OS cells were transfected with pMT2 (together with β-gal to identify the transfected cells), pMT2-HA-WT-FAST or pMT2-HA- Y428G-FAST. After 48 h, cells were further cultured in the absence or presence of anti-Fas for 6 h, then processed for immunofluorescence to quantify the percentage of transfected cells in which caspase-3 has been activated. In HeLa cells, WT-FAST, but not Y428G-FAST, inhibited the Fas-induced activation of caspase-3 (). In U2OS cells, FAST also inhibited the Fas-induced activation of caspase-3 (). In contrast, Y428G-FAST significantly enhances Fas-induced activation of caspase-3, suggesting that it can act as a dominant negative inhibitor of endogenous FAST in these cells.
Figure 9.WT FAST, but not its Y428G mutant, inhibits Fas-induced activation of caspase-3. HeLa cells (A) and U2OS cells (B) were co-transfected with β-gal together with vector control, WT-FAST, or Y428G-FAST. After 48 h, cells were processed for immunofluorescence microscopy to quantify the expression of active caspase-3 in β-gal transfectants.
Discussion
TIA-1 is an RNA-binding protein that recognizes adenine/uridine-rich elements such as AUUUA sequences in the 3′-untranslated regions of selected mRNAs.Citation1 TIA-1 inhibits the translation of these transcripts by promoting the assembly of a stalled pre-initiation complex.Citation2 TIA-1 also is a component of SGs which are discrete cytoplasmic foci at which untranslated mRNAs accumulate in cells exposed to environmental stress.Citation2 These functional attributes are similar to those of CPEB and Bruno, which are proteins that bind to regulatory elements found in the 3′-untranslated regions of selected mRNAs to promote translational silencing and assembly of cytoplasmic mRNP granules.Citation6,Citation10 Like TIA-1, CPEB is a component of stress granules.Citation6 Both CPEB and Bruno interact with eIF4E-binding proteins (maskin and cup, respectively) that inhibit translation by displacing eIF4G from the initiation complex.Citation10 FAST is a TIA-1-interacting protein whose overexpression antagonizes the functional effects of TIA-1.Citation9 The possibility that FAST may be a functional ortholog of maskin and cup was suggested by the identification of an eIF4E-binding motif 428YXXXXLΦ433 within the coding sequence of FAST. This motif conforms to the consensus interaction domain that allows eIF4GI, eIF4GII, 4E-BP1, 4E-BP2, 4E-BP3, 4E-T and cup to bind to eIF4E.Citation11,Citation14,Citation15,Citation19 Immunoprecipitation analysis confirmed that FAST binds to eIF4E. A Y428G mutation that disrupts the second consensus binding motif does not bind to eIF4E, indicating that this motif is required for FAST:eIF4E interactions. It remains to be determined whether the first eIF4E binding motif is involved in FAST:eIF4E interactions.
Whereas overexpression of TIA-1 inhibits the expression of co-transfected β-gal, overexpression of FAST, but not FAST (Y428G), enhances the expression of co-transfected β-gal. This result indicates that FAST:eIF4E interactions are required for the enhanced expression of β-gal. It is possible that overexpressed recombinant FAST dominantly inhibits the assembly of TIA-1:FAST:eIF4E inhibitory complexes to allow increased translation of β-gal. However, the ability of FAST, but not FAST (Y428G), to stabilize β-gal mRNA indicates that FAST:eIF4E interactions can also regulate mRNA degradation. Recent observations relating to the cell biology of mRNA metabolism may be relevant to the TIA-1:FAST:eIF4E regulatory pathway. Selected mRNAs accumulate at discrete cytoplasmic foci that remodel and/or determine the fate of mRNP complexes. Stress granules (SGs) are cytoplasmic aggregates of stalled preinitiation complexes that accumulate during stress.Citation1 Processing bodies (PBs) are distinct cytoplasmic sites of mRNA degradation.Citation20-Citation26 Vital microscopy has shown that PBs intermittently dock at SGs revealing dynamic contacts between these structures.Citation5 The ability of the mRNA decay factors TTP and BRF-1 to induce fusion between SGs and PBs and direct ARE-containing transcripts to PBsCitation27 suggests that mRNA released from disassembled polysomes is sorted and remodeled at SGs, from which selected transcripts are delivered to PBs for degradation.Citation5 Interestingly, FAST and eIF4E are components of both SGs and PBs.Citation7 In contrast, eIF4G is restricted to SGs.Citation7 If it is true that mRNA moves from SGs to PBs, these results suggest that eIF4G is displaced from mRNP complexes before they are delivered to the PB. It is therefore possible that interactions between FAST, TIA-1, and eIF4E influence the composition and/or transport of mRNPs as they move from SGs to PBs. Like FAST, eIF4E-T is localized at P bodies.Citation28 Thus, eIF4E-binding proteins may play an important role in regulating the decapping reaction at the P body. Additional experiments will be required to sort out the molecular details by which FAST, TIA-1 and eIF4E regulate mRNA metabolism.
We previously reported that overexpressed recombinant FAST inhibits Fas-induced activation of caspase-3.Citation9 This functional effect appears to be due to enhanced expression of inhibitors of apoptosis such as c-IAP-1 and XIAP. In contrast to WT FAST, its Y428G mutant does not inhibit Fas-induced activation of caspase-3. This finding implicates FAST:eIF4E complexes in this functional response. It remains to be determined if increased expression of IAP proteins results from enhanced mRNA stability. In U2OS cells, the Y428G mutant of FAST significantly enhances Fas-induced apoptosis. This result suggests that the mutant can function as a dominant negative inhibitor of endogenous FAST in these cells.
Our results indicate that interactions between FAST and eIF4E can stabilize mRNA and inhibit apoptosis. We previously reported that TIA-1 antagonizes the functional effects of FAST.Citation9 Increased levels of TIA-1 in cells prevents FAST from promoting the expression of β-gal and inhibiting Fas-induced apoptosis.Citation9 Taken together, these results suggest that the assembly of TIA-1:FAST:eIF4E complexes can regulate mRNA stability, protein translation and cell survival. This complex may be functionally analogous to the CPEB:maskin:eIF4E and Bruno:Cup:eIF4E complexes that regulate the expression of selected proteins during oogenesis.Citation10
Materials and Methods
Cells, Antibodies (Abs), and Reagents
COS-7 and HeLa cells were obtained from the American Type Culture Collection. U2OS cells were a gift from Dr. John Blenis (Harvard Medical School, Boston, MA). Cells were maintained in 10% FCS in DMEM.
Antibodies obtained from commercial sources include anti-HA (monoclonal Ab clone 16B12, IgG1; Berkeley Ab Co.), anti-eIF4E (rabbit polyclonal Ab, Cell Signaling), anti-eIF4G (rabbit polyclonal Ab, Santa Cruz), anti-FLAG (murine monoclonal Ab M2 clone, Sigma), anti-active caspase 3 (rabbit polyclonal Ab, Promega), anti-β-gal (monoclonal Ab (Promega), anti-myc (rabbit polyclonal Ab, Santa Cruz). Isotype-specific secondary Abs (ML grade) (Jackson ImmunoResearch Labs). Hoechst dye 33258 and other chemicals (Sigma Chemical Co). m7GTP-Sepharose was from GE Healthcare.
Plasmid constructions
Full length recombinant FAST was subcloned into the pMT2-HA vector and pcDNA3-FLAG as described previously.Citation8 The FAST mutant that substitutes G at Y428 in the eIF4E-binding motif was constructed using PCR mediated mutagenesis with primers 5′-GACCTGACTGTGCCTCCAGGCGGCTGCACAGACTTCCTGCTG-3′ and 5′-CAGCAGGAAGTCTGTGCAGCCGCCTGGAGGCACAGTCAGGTC-3′. pcDNA3-FLAG-eIF4E is described elsewhere (Kedersha et al., submitted). For pMT2-HA-eIF4E, eIF4E was amplified from plasmid pcDNA3-eIF4E (a gift from Dr. Dan Dixon, Univ. South Carolina, Columbia, SC) using primers 5′-CACAGAATTCTTAGTGTGGAGCCGCTCTTAG-3′ and 5′-CACGAATTCGCGACTGTCGAACCGGAAACC-3′, digested with EcoRI, and ligated into the EcoRI sites of the vector pMT2-HA. The sequences of all constructs were verified before use. pcDNA3-β-gal was obtained from Invitrogen. pcDNA3–6myc-cellular inhibitor of apoptosis-1 (cIAP-1) was a gift from Dr. Martin Holcik (CHEO Research Institute).
SDS-PAGE immunoblot analysis
Recombinant proteins were boiled in SDS sample buffer, resolved on 4–20% polyacrylamide gradient gels (Invitrogen Corp.), and transferred to nitrocellulose. Whole cells were solubilized in SDS sample buffer, then boiled and sonicated to shear DNA. Whole cell extracts were then separated on 4–20% gradient gels, transferred to nitrocellulose, and probed with the indicated antibodies using previously described methods.Citation8
Transfections
Cells were transfected using Superfect (Qiagen) or Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Immunoprecipitations were performed as previously described.Citation29 For each ip, anti-HA ascites fluid was used at 1/1000.
RNA gel blot analysis
Total RNA was extracted using RNAqueous (Ambion) according to the manufacturer’s instructions. RNA (3–10 μg) was resolved by 1% agarose/2% formaldehyde MOPS gel electrophoresis, blotted onto Nytran Supercharge membranes (Schleicher and Schuell) using 8x SSC and hybridized overnight at 50°C with digoxigenin-labeled DNA probes in DIG Easy Hyb solution (Roche). After washing at 50°C with 2x SSC/0.1% SDS twice and 0.5x SSC/0.1% SDS twice (20 min each), the membranes were placed in Blocking Reagent (Roche) for 30 min at room temperature, probed with alkaline phosphatase-labeled anti-digoxigenin Ab (Roche) for 30 min, and washed for 30–60 min with Dig Wash Buffer (100 mM NaCl, 0.3% Tween-20, and 130 mM TRIS-HCl, pH 7.5,). Signals were visualized with CDP-Star (Roche). Probes were generated by PCR using digoxigenin-labeled nucleotides (Roche), β-gal cDNA or glyceraldehyde 3-phosphate dehydrogenase (GAPDH) cDNA and the primer pairs 5′-β-gal/3′-β-gal or 5′-GAPDH/3′-GAPDH, respectively. The sequence for primer 5′ β-gal is 5′-CAGCTGGCGCAGGTAGCA-3′, the sequence for primer 3′ β-gal is 5′-CCGCCGATACTGACGGGC-3′, the sequence for 5′-GAPDH is 5′-TCCTGCACCACCAACTGCTTAGC-3′, the sequence for 3′-GAPDH is 5′-TGATGTCATCATACTTGGCAG-3′. For mRNA stability analysis, cells were first treated with 5 mg/ml of actinomycin D for various times before processed for RNA gel blot analysis.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank members of the Anderson lab for helpful suggestions and discussions. We thank Ms. Natalie Gilks for making the pMT2-HA-eIF4E construct, and Dr. Nancy Kedersha for critical review of the manuscript. This work was supported by NIH grants AI059746, AI050167, AI033600, and AI065858.
REFERENCES
- Anderson P, Kedersha N. Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones 2002; 7:213 - 21; http://dx.doi.org/10.1379/1466-1268(2002)007<0213:VSTROE>2.0.CO;2; PMID: 12380690
- Anderson P, Kedersha N. Stressful initiations. J Cell Sci 2002; 115:3227 - 34; PMID: 12140254
- Dember LM, Kim ND, Liu KQ, Anderson P. Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J Biol Chem 1996; 271:2783 - 8; http://dx.doi.org/10.1074/jbc.271.5.2783; PMID: 8576255
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol 1999; 147:1431 - 42; http://dx.doi.org/10.1083/jcb.147.7.1431; PMID: 10613902
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol 2005; 169:871 - 84; http://dx.doi.org/10.1083/jcb.200502088; PMID: 15967811
- Wilczynska A, Aigueperse C, Kress M, Dautry F, Weil D. The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J Cell Sci 2005; 118:981 - 92; http://dx.doi.org/10.1242/jcs.01692; PMID: 15731006
- Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci 2008; 33:141 - 50; http://dx.doi.org/10.1016/j.tibs.2007.12.003; PMID: 18291657
- Tian Q, Taupin J, Elledge S, Robertson M, Anderson P. Fas-activated serine/threonine kinase (FAST) phosphorylates TIA-1 during Fas-mediated apoptosis. J Exp Med 1995; 182:865 - 74; http://dx.doi.org/10.1084/jem.182.3.865; PMID: 7544399
- Li W, Simarro M, Kedersha N, Anderson P. FAST is a survival protein that senses mitochondrial stress and modulates TIA-1-regulated changes in protein expression. Mol Cell Biol 2004; 24:10718 - 32; http://dx.doi.org/10.1128/MCB.24.24.10718-10732.2004; PMID: 15572676
- Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 2005; 433:477 - 80; http://dx.doi.org/10.1038/nature03205; PMID: 15690031
- Mader S, Lee H, Pause A, Sonenberg N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol Cell Biol 1995; 15:4990 - 7; PMID: 7651417
- Pause A, Belsham GJ, Gingras AC, Donzé O, Lin TA, Lawrence JC Jr., et al. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature 1994; 371:762 - 7; http://dx.doi.org/10.1038/371762a0; PMID: 7935836
- Poulin F, Gingras AC, Olsen H, Chevalier S, Sonenberg N. 4E-BP3, a new member of the eukaryotic initiation factor 4E-binding protein family. J Biol Chem 1998; 273:14002 - 7; http://dx.doi.org/10.1074/jbc.273.22.14002; PMID: 9593750
- Dostie J, Ferraiuolo M, Pause A, Adam SA, Sonenberg N. A novel shuttling protein, 4E-T, mediates the nuclear import of the mRNA 5′ cap-binding protein, eIF4E. EMBO J 2000; 19:3142 - 56; http://dx.doi.org/10.1093/emboj/19.12.3142; PMID: 10856257
- Gradi A, Imataka H, Svitkin YV, Rom E, Raught B, Morino S, et al. A novel functional human eukaryotic translation initiation factor 4G. Mol Cell Biol 1998; 18:334 - 42; PMID: 9418880
- Nelson MR, Leidal AM, Smibert CA. Drosophila Cup is an eIF4E-binding protein that functions in Smaug-mediated translational repression. EMBO J 2004; 23:150 - 9; http://dx.doi.org/10.1038/sj.emboj.7600026; PMID: 14685270
- von der Haar T, Gross JD, Wagner G, McCarthy JE. The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat Struct Mol Biol 2004; 11:503 - 11; http://dx.doi.org/10.1038/nsmb779; PMID: 15164008
- Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol 2004; 11:121 - 7; http://dx.doi.org/10.1038/nsmb724; PMID: 14749774
- Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol Cell 1999; 3:707 - 16; http://dx.doi.org/10.1016/S1097-2765(01)80003-4; PMID: 10394359
- Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 2003; 300:805 - 8; http://dx.doi.org/10.1126/science.1082320; PMID: 12730603
- Cougot N, van Dijk E, Babajko S, Séraphin B. ‘Cap-tabolism’. Trends Biochem Sci 2004; 29:436 - 44; http://dx.doi.org/10.1016/j.tibs.2004.06.008; PMID: 15362228
- Cougot N, Babajko S, Séraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol 2004; 165:31 - 40; http://dx.doi.org/10.1083/jcb.200309008; PMID: 15067023
- Eystathioy T, Chan EK, Tenenbaum SA, Keene JD, Griffith K, Fritzler MJ. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol Biol Cell 2002; 13:1338 - 51; http://dx.doi.org/10.1091/mbc.01-11-0544; PMID: 11950943
- Eystathioy T, Jakymiw A, Chan EK, Séraphin B, Cougot N, Fritzler MJ. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA 2003; 9:1171 - 3; http://dx.doi.org/10.1261/rna.5810203; PMID: 13130130
- Ingelfinger D, Arndt-Jovin DJ, Lührmann R, Achsel T. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA 2002; 8:1489 - 501; PMID: 12515382
- Yang Z, Jakymiw A, Wood MR, Eystathioy T, Rubin RL, Fritzler MJ, et al. GW182 is critical for the stability of GW bodies expressed during the cell cycle and cell proliferation. J Cell Sci 2004; 117:5567 - 78; http://dx.doi.org/10.1242/jcs.01477; PMID: 15494374
- Franks TM, Lykke-Andersen J. TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes Dev 2007; 21:719 - 35; http://dx.doi.org/10.1101/gad.1494707; PMID: 17369404
- Andrei MA, Ingelfinger D, Heintzmann R, Achsel T, Rivera-Pomar R, Lührmann R. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA 2005; 11:717 - 27; http://dx.doi.org/10.1261/rna.2340405; PMID: 15840819
- Li W, Kedersha N, Chen S, Gilks N, Lee G, Anderson P. FAST is a BCL-X(L)-associated mitochondrial protein. Biochem Biophys Res Commun 2004; 318:95 - 102; http://dx.doi.org/10.1016/j.bbrc.2004.03.188; PMID: 15110758