Abstract
The ATP-dependent RNA helicase UPF1, a key factor in nonsense-mediated mRNA decay (NMD), was so far thought to be recruited specifically to NMD-targeted mRNAs by aberrantly terminating ribosomes. However, two recent publications reporting independently transcriptome-wide mapping of UPF1 occupancy on RNA challenge this model and instead provide evidence that UPF1 binds to mRNA already before translation. According to the new data, UPF1 appears to initially bind all mRNAs along their entire length and gets subsequently stripped off the coding sequence by translating ribosomes. This re-poses the question of where and how UPF1 engages with mRNA and how the NMD-targeted transcripts are selected among the UPF1-bound mRNAs.
Nonsense-Mediated mRNA Decay
During their entire lifespan, mRNAs are accompanied by a changing set of proteins and small non-coding (nc)RNAs forming complex messenger riboncleoprotein particles (mRNPs).Citation1 The components of an mRNP and their positioning on the mRNA to a large extent determine the fate of the mRNA molecule and hence play important roles in the regulation of gene expression. Cells possess multiple surveillance mechanisms that detect aberrant mRNPs and promote the degradation of the concerned mRNA. Arguably the best-studied surveillance mechanism is nonsense-mediated mRNA decay (NMD), which was discovered more than 30 y ago and initially described as a translation-dependent process that selectively degrades mRNAs with truncated open reading frames (ORFs) due to premature translation-termination codons (PTC),Citation2,Citation3 thereby preventing the expression of potentially deleterious C-terminally truncated proteins. In humans, one out of three disease-associated mutations leads to the production of PTC-containing mRNA and NMD therefore influences the severity of the clinical manifestations caused by these mutations.Citation4-Citation6 Besides its quality control function, genome-wide mRNA profiling experiments revealed that NMD also regulates the abundance of many physiological mRNAs coding for functional proteins (reviewed inCitation7).
Unraveling UPF1 Functions—The Key to Understand NMD
In spite of intensive research over the past decades, our understanding of the molecular mechanism of NMD is still fragmentary. Among the so far identified proteins involved in NMD, UPF1 plays a – if not the – key role: with 48.5% amino acid identity between human and baker’s yeast it is the most conserved NMD factor,Citation8 it is essential in vertebrates and flies,Citation9-Citation11 and binds RNA unlike most other NMD factors. With an estimated 3*106 molecules per cell, UPF1 is a quite abundant protein.Citation12 In relation to the approx. 3*105 mRNA molecules per cell,Citation13 there are on average about 10 UPF1 proteins available per mRNA, which is comparable to the approx. Twenty molecules of poly(A) binding protein (PABP) per mRNA.Citation14
UPF1 is an ATP-dependent 5′-3′ RNA helicase belonging to the superfamily 1 (SF1).Citation15 Two RecA-like domains form the helicase domain with the ATP-binding pocket located in between.Citation16 The helicase and the ATPase activities of UPF1 are repressed by an N-terminal cysteine/histidine-rich zinc finger (CH) domain and a C-terminal domain rich in serine-glutamine motifs (SQ domain).Citation17-Citation19 Since UPF1’s ATPase activity is required for NMD, the inhibitory configurations of the flanking CH and SQ domains must be relieved at some point during the NMD process. While the inhibitory effect of the CH domain is suspended by its interaction with UPF2, which pulls the CH domain from the Rec2A subdomain to the Rec1A subdomain on the opposite side of the helicase core,Citation17 it is currently unknown how the SQ domain-mediated inhibition is overcome.Citation19 There is evidence that the ATPase activity of UPF1 is required for disassembling the NMD complex after the endonucleolytic cleavage of the target mRNA, since UPF1 mutants unable to bind or hydrolyze ATP led to the accumulation of 3′ RNA decay intermediates.Citation20 As expected for a helicase, UPF1 has the capacity to unwind RNA duplexes in vitro, an activity that is promoted by the other two conserved NMD factors UPF2 and UPF3B,Citation18 but it remains to be seen if UPF1 also translocates on RNA in vivo.
In metazoans, UPF1 is a phosphoprotein with more than a dozen serine or threonine followed by glutamine (S/TQ) motifs, and half of them can be phosphorylated in vitro by the phosphatidylinositol 3-kinase-related protein kinase SMG1. In vivo, UPF1 undergoes a cycle of phosphorylation and dephosphorylation, which is essential for NMD in metazoans (reviewed inCitation21). The exact function of UPF1 phosphorylation during NMD and its relation to ATP binding and hydrolysis is not known, but the endonuclease SMG6 and the decay factor-adaptors SMG5/SMG7 and PNRC2 preferentially interact with phosphorylated UPF1 and ATPase-deficient UPF1 mutants accumulate in a hyper-phosphorylated state.Citation22-Citation24 Recently, Yamashita and colleagues identified phospho-T28 and phospho-S1096 to be required for the interaction of UPF1 with SMG6 and SMG7, respectively.Citation25
Current Model: Selective and Translation-Dependent Recruitment of UPF1 to NMD-Targeted mRNAs
A central unsolved question is how the NMD pathway selects its target mRNAs. Based on a wealth of biochemical and genetic data, including the above-mentioned characteristics of UPF1, the following working model has emerged during the last years: in a nutshell, NMD ensues on mRNAs with termination codons in an environment unfavorable for efficient translation termination.Citation7,Citation26 The underlying assumption is that the translation termination is a highly regulated process involving the orchestrated function of many factors, comparable to translation initiation, and that in the absence of a termination promoting factor, ribosomes stall at the termination codonCitation27,Citation28 and lead to the assembly of NMD factors that ultimately degrade the mRNA. Supporting this model, UPF1 was found to interact with the release factors eRF1 and eRF3Citation23,Citation29,Citation30 and this interaction is antagonized by the interaction between eRF3 and poly(A) binding protein C1 (PABPC1).Citation31 PABPC1 and its yeast homolog Pab1p have been shown to stimulate terminationCitation32-Citation34 and tethering of PABPC1/Pab1p near NMD-triggering termination codons efficiently suppresses NMD.Citation27,Citation31,Citation34-Citation36 This model provides an explanation why not only PTCs but also long 3′ UTRs can elicit NMD.Citation36-Citation38 The detection of an immunoprecipitable complex consisting of SMG1, UPF1, eRF1 and eRF3 (called SURF complexCitation23) suggested that SURF might form at ribosomes that fail to terminate properly. According to this model, UPF1 would be recruited in a translation-dependent manner and only to NMD-targeted transcripts. This view was corroborated by studies reporting preferential association of UPF1 with PTC-containing transcripts in C. elegansCitation39 and in human cells.Citation40
In the SURF complex, SMG1 kinase is kept in an inactive state by two regulatory proteins, SMG8 and SMG9, until UPF2 interacts with UPF1.Citation41 The conformational change induced by UPF2 leads to the dissociation of SMG8 and SMG9 and UPF1 phosphorylation by the activated SMG1 kinaseCitation41,Citation25. As already mentioned above, UPF1 phosphorylation is then thought to commit the mRNA to degradation by the recruitment of the decay promoting factors SMG6, SMG5/SMG7 and PNRC2 (reviewed inCitation7).
In the case of exon junction complex (EJC)-independent NMD, free UPF2 would have to join the SURF complex by diffusion, which is expected to be a rate-limiting step and results in rather inefficient NMD.Citation37,Citation42 In contrast, classical NMD-targeted mRNAs harbor an NMD-enhancing EJC downstream of the termination codon. The tetrameric EJC core consisting of eIF4AIII, MAGO, MLN51 and Y14 binds to UPF3, which in turn recruits UPF2.Citation43-Citation46 EJCs assemble at exon-exon junctions during splicingCitation47-Citation50 and are displaced from the coding sequence during translation by the scanning ribosomes,Citation51,Citation52 so that after onset of translation most mRNAs will be devoid of EJCs. However, if the termination codon is located more than 30 nucleotides upstream of the 3′-most EJC, which often is the case for PTCs, these EJCs remain bound to the mRNA and enhance NMD by providing pre-bound UPF2 ready to interact with the SURF complex (reviewed inCitation7).
Evidence for Translation-Independent mRNA Interaction of UPF1
The recent development of CLIP-seq (crosslinking and immunoprecipitation followed by high-throughput sequencing) techniques allows the transcriptome-wide detection of the binding sites for RNA-binding proteins.Citation53,Citation54 To explore how NMD shapes the embryonic transcriptome, Hurt and colleagues performed integrated genome-wide analyses of UPF1 binding locations, NMD-regulated gene expression, and translation in murine embryonic stem cells (mESCs),Citation55 and we used CLIP-seq to map the RNA-binding sites of UPF1 in human cells.Citation56 In both cell types, UPF1 was detected predominantly in 3′ UTR sequences, where it was quite evenly distributed, whereas coding sequence (CDS) was much less covered with UPF1. This high enrichment of UPF1 in 3′ UTRs is consistent with a study from Hogg and Goff, who investigated UPF1 association with PP7-tagged reporter transcripts and found that the amount of UPF1 interacting with the transcripts correlated with their 3′ UTR length.Citation57 Intriguingly and contradicting the above-described NMD model, Hogg and Goff further reported that the UPF1 interaction with the tested 3′ UTRs occurred even in the absence of translation. To explore this discrepancy, both CLIP-seq studies also analyzed RNA binding of UPF1 in translation-suppressed HeLa cellsCitation56 and mESCs,Citation55 respectively. Translation inhibition unveiled a striking redistribution of UPF1 toward CDS,Citation55,Citation56 indicating that before translation UPF1 interacts with mRNAs along its entire length and that during translation elongating ribosomes displace it from the CDS (). Displacement of UPF1 from RNA by elongating ribosomes is consistent with results from Hogg and Goff, who showed that even a low frequency of translational read-through induced by retroviral RNA elements preceding the stop codon reduced the UPF1 association with long 3′ UTRs and stabilized the otherwise NMD-targeted reporter transcripts.Citation57 Since CLIP yields relative data, i.e., enrichment of sequence x in the IP relative to its abundance in the transcriptome, the shift of UPF1 occupancy from 3′ UTR to CDS upon translation inhibition could result from an increased binding to CDS, a decreased binding to 3′ UTRs or a combination of both. To distinguish between these possibilities, we performed RNA-immunoprecipitations (RIP) in which CDS and 3′ UTR of selected endogenous mRNAs were separated by an oligo-mediated RNase H cleavage during the IP.Citation56 This assay confirmed the marked increase of UPF1 association with CDS upon translation inhibition, corroborating the conclusion that UPF1 binds to CDS in the absence of translation and gets displaced from CDS during translation.
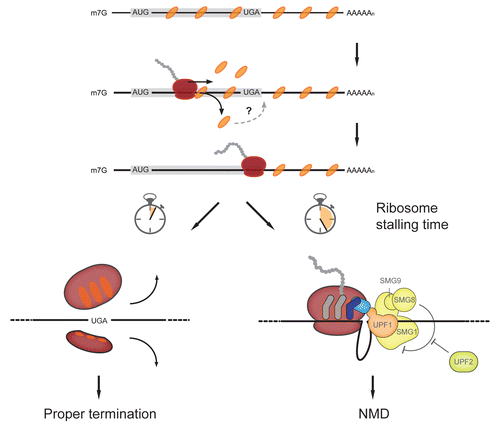
Figure 1. Modified working model for NMD based on the new evidence for UPF1 interaction with mRNA before translation. The coding sequence (CDS), starting with AUG and ending with a stop codon (UGA in this Figure), is depicted by a light gray box. UPF1 (orange ovals) associates along the entire mRNA during or after splicing, but before translation starts. The ribosome (red) then displaces UPF1 from the CDS. It is not known whether displaced UPF1 can rebind in the UTR regions (indicated by the question mark). According to the model, the kinetics of translation termination determines if NMD ensues. Proper termination is typified by a short residence time of the ribosme at the stop codon (bottom left), while the absence of termination promoting signals (e.g., PABPC1) stalls the ribosome at the stop codon, allowing remaining UPF1 to interact with the release factors (eRFs, light blue) and the SMG1 complex (yellow; bottom right). Interaction of UPF2 (light green) with the UPF1:SMG1:eRFs (SURF) complex induces a conformational change that leads to dissociation of SMG8 and SMG9 and to the activation of SMG1’s kinase activity. Phosphorylated UPF1 subsequently recruits directly (SMG6) and/or indirectly (SMG5/SMG7) RNA decay factors, leading to the degradation of the mRNA.
Furthermore, association of UPF1 with long ncRNAs was also detected, providing additional evidence for translation-independent binding.Citation56 As expected for untranslated RNA, the UPF1 binding on these transcripts was not altered by translation inhibition.
Specificity of UPF1 for mRNA
What do the two CLIP papers tell us about the specificity of the UPF1-RNA interaction? As one might have expected for a helicase, motif searches did not reveal any preferred RNA binding sequence for UPF1. Nevertheless, UPF1 was clearly distributed along RNA in a non-random fashion. Hurt and colleagues found an enriched UPF1 density in G-rich areas with an increased propensity to form secondary structures.Citation55 Our analysis did not reveal any sequence preference except for a bias toward U at the position of the crosslink,Citation56 which most likely just reflects the better UV crosslinking efficiency of U compared with the other bases.Citation58
Interestingly with regards to the question of when and how UPF1 is recruited to RNA, UPF1 density was 10 – 30-fold higher in exons than introns in both studies, and also very little UPF1 association was detected with rRNA and tRNA. This suggests that UPF1 associates with mRNA during or after splicing. UPF1 recruitment to mRNA during splicing together with the EJC seemed to be an attractive hypothesis at first glance, because UPF1 interacts with the EJC through UPF2 and UPF3,Citation46 but both CLIP studies did find no correlation between UPF1 binding and EJC deposition. Hurt and colleagues showed that genes encoding for transcripts with high UPF1 binding density in their 3′ UTR were not enriched for the expression of downstream EJC-containing isoforms,Citation55 and we did not find any correlation between the UPF1 and the recently reported EJC binding sites.Citation56,Citation59 Moreover, knockdown of the EJC core factor eIF4AIII did not affect UPF1 association with mRNA.Citation60 Thus, the available data indicates that UPF1 engages with mRNA independently of EJCs and the specificity of UPF1 for mRNA remains to be explored in the future. It is conceivable that UPF1 binds RNA in the cytoplasm and that mRNA specificity results from the fact that the highly structured tRNAs and the rRNAs, which assemble through a highly orchestrated process into ribosomal subunits, are simply not accessible for UPF1 binding.
If not UPF1 Binding, What is the Distinctive Step in NMD Target Selection?
According to the above-described NMD model, UPF1 recruitment to an mRNA marks this mRNA for NMD and UPF1 should therefore preferentially – if not exclusively – be associated with NMD targeted mRNAs. Consistent with this model, a recent study conducted with a set of reporter transcripts reported the accumulation of more UPF1 molecules per 3′ UTR length unit if the mRNA was an NMD target.Citation40 However, the transcriptome-wide analyses gave a more complex picture: the results of Hurt and colleagues indicate a correlation between UPF1 binding and NMD only for transcripts with short 3′ UTRs, most of which harbor an EJC as the NMD-inducing signal.Citation55 However, no significant difference in UPF1 density was detected when we compared two sets of experimentally identified endogenous NMD targetsCitation38,Citation61 with corresponding control groups of transcripts that are not affected by NMD.Citation56
The lack of a strict correlation between UPF1 binding and NMD together with the evidence for translation-independent binding of UPF1 to mRNA challenges the idea that UPF1 binding would be the step that commits an mRNA to the NMD pathway. At the same time, it re-opens the question about what the NMD-triggering step then would be. At this moment, one can only speculate and more research is clearly needed. An obvious possibility is that the SMG1-mediated phosphorylation of UPF1, which leads to recruitment of RNA degradation factors (see above), represents the activation step of NMD that specifically occurs when ribosomes fail to properly terminate translation (). A variation of the same theme is that UPF1 indiscriminately binds to mRNA and acts there like a ticking bomb: if not stripped off by ribosomes within a certain time window, it will become phosphorylated and induce the decay of the mRNA. Both scenarios predict that hyper-phosphorylated UPF1 would be found associated specifically with NMD targeted transcripts, a prediction that can be tested by purification of affinity-tagged reporter transcripts. It could also be that ATP binding or hydrolysis by UPF1 are the crucial steps to activate NMD.
Crosslink Sites do not Necessarily Represent the Locations of Initial Binding
Another open question is whether UPF1 initially engages with mRNA at specific sites. The crosslinking sites detected in the two CLIP studies do not necessarily represent the positions of initial UPF1 binding to mRNA, because UPF1 can probably translocate on RNA by virtue of its 5′-to-3′ helicase activity. An interesting follow-up study would therefore be to CLIP exogenously expressed tagged UPF1 mutants defective in ATP binding, ATP hydrolysis and/or helicase activity.
UPF1 Reveals its Secrets only Slowly
While it is obvious to all researchers in the field that understanding UPF1 function is key to understand NMD, this protein does not divulge its secrets easily. Our current understanding can be summarized by the following statement by J.R. Hogg: “…UPF1 accumulation in mRNPs is a prerequisite for decay but is not sufficient to consign the mRNP to destruction. Instead the initiation of decay requires the completion of one or more additional rate-limiting steps…”Citation62. Additional investigations of the activities, regulation and interactions of UPF1 and other NMD proteins in the context of endogenous mRNPs are needed to reveal the order of events leading from UPF1 accumulation in mRNPs to the decay of the mRNAs.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclose
Acknowledgments
The NMD research in the laboratory is funded by grants to O.M. from the European Research Council (ERC-StG 207419), the Swiss National Science Foundation (31003A-127614 and 31003A-143717), the Novartis Foundation for Biomedical Research, and the canton Bern.
References
- Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science 2005; 309:1514 - 8; PMID: 16141059
- Losson R, Lacroute F. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc Natl Acad Sci U S A 1979; 76:5134 - 7; PMID: 388431
- Maquat LE, Kinniburgh AJ, Rachmilewitz EA, Ross J. Unstable beta-globin mRNA in mRNA-deficient beta o thalassemia. Cell 1981; 27:543 - 53; PMID: 6101206
- Bhuvanagiri M, Schlitter AM, Hentze MW, Kulozik AE. NMD: RNA biology meets human genetic medicine. Biochem J 2010; 430:365 - 77; PMID: 20795950
- Khajavi M, Inoue K, Lupski JR. Nonsense-mediated mRNA decay modulates clinical outcome of genetic disease. Eur J Hum Genet 2006; 14:1074 - 81; PMID: 16757948
- Kuzmiak HA, Maquat LE. Applying nonsense-mediated mRNA decay research to the clinic: progress and challenges. Trends Mol Med 2006; 12:306 - 16; PMID: 16782405
- Schweingruber C, Rufener SC, Zünd D, Yamashita A, Mühlemann O. Nonsense-mediated mRNA decay - mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim Biophys Acta 2013; 1829:612 - 23; PMID: 23435113
- Culbertson MR, Leeds PF. Looking at mRNA decay pathways through the window of molecular evolution. Curr Opin Genet Dev 2003; 13:207 - 14; PMID: 12672499
- Avery P, Vicente-Crespo M, Francis D, Nashchekina O, Alonso CR, Palacios IM. Drosophila Upf1 and Upf2 loss of function inhibits cell growth and causes animal death in a Upf3-independent manner. RNA 2011; 17:624 - 38; PMID: 21317294
- Medghalchi SM, Frischmeyer PA, Mendell JT, Kelly AG, Lawler AM, Dietz HC. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum Mol Genet 2001; 10:99 - 105; PMID: 11152657
- Wittkopp N, Huntzinger E, Weiler C, Saulière J, Schmidt S, Sonawane M, Izaurralde E. Nonsense-mediated mRNA decay effectors are essential for zebrafish embryonic development and survival. Mol Cell Biol 2009; 29:3517 - 28; PMID: 19414594
- Pal M, Ishigaki Y, Nagy E, Maquat LE. Evidence that phosphorylation of human Upfl protein varies with intracellular location and is mediated by a wortmannin-sensitive and rapamycin-sensitive PI 3-kinase-related kinase signaling pathway. RNA 2001; 7:5 - 15; PMID: 11214180
- Velculescu VE, Madden SL, Zhang L, Lash AE, Yu J, Rago C, Lal A, Wang CJ, Beaudry GA, Ciriello KM, et al. Analysis of human transcriptomes. Nat Genet 1999; 23:387 - 8; PMID: 10581018
- Görlach M, Burd CG, Dreyfuss G. The mRNA poly(A)-binding protein: localization, abundance, and RNA-binding specificity. Exp Cell Res 1994; 211:400 - 7; PMID: 7908267
- Jankowsky E. RNA helicases at work: binding and rearranging. Trends Biochem Sci 2011; 36:19 - 29; PMID: 20813532
- Cheng Z, Muhlrad D, Lim MK, Parker R, Song H. Structural and functional insights into the human Upf1 helicase core. EMBO J 2007; 26:253 - 64; PMID: 17159905
- Chakrabarti S, Jayachandran U, Bonneau F, Fiorini F, Basquin C, Domcke S, Le Hir H, Conti E. Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Mol Cell 2011; 41:693 - 703; PMID: 21419344
- Chamieh H, Ballut L, Bonneau F, Le Hir H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol 2008; 15:85 - 93; PMID: 18066079
- Fiorini F, Boudvillain M, Le Hir H. Tight intramolecular regulation of the human Upf1 helicase by its N- and C-terminal domains. Nucleic Acids Res 2012; advanced online publication, Dec. 28, 2012.
- Franks TM, Singh G, Lykke-Andersen J. Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense- mediated mRNA decay. Cell 2010; 143:938 - 50; PMID: 21145460
- Yamashita A. Role of SMG-1-mediated Upf1 phosphorylation in mammalian nonsense-mediated mRNA decay. Genes Cells 2013; 18:161 - 75; PMID: 23356578
- Cho H, Kim KM, Kim YK. Human proline-rich nuclear receptor coregulatory protein 2 mediates an interaction between mRNA surveillance machinery and decapping complex. Mol Cell 2009; 33:75 - 86; PMID: 19150429
- Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev 2006; 20:355 - 67; PMID: 16452507
- Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JW, Maquat LE. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell 2008; 133:314 - 27; PMID: 18423202
- Okada-Katsuhata Y, Yamashita A, Kutsuzawa K, Izumi N, Hirahara F, Ohno S. N- and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Res 2012; 40:1251 - 66; PMID: 21965535
- Kervestin S, Jacobson A. NMD: a multifaceted response to premature translational termination. Nat Rev Mol Cell Biol 2012; 13:700 - 12; PMID: 23072888
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 2004; 432:112 - 8; PMID: 15525991
- Peixeiro I, Inácio A, Barbosa C, Silva AL, Liebhaber SA, Romão L. Interaction of PABPC1 with the translation initiation complex is critical to the NMD resistance of AUG-proximal nonsense mutations. Nucleic Acids Res 2012; 40:1160 - 73; PMID: 21989405
- Czaplinski K, Ruiz-Echevarria MJ, Paushkin SV, Han X, Weng Y, Perlick HA, Dietz HC, Ter-Avanesyan MD, Peltz SW. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev 1998; 12:1665 - 77; PMID: 9620853
- Wang W, Czaplinski K, Rao Y, Peltz SW. The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J 2001; 20:880 - 90; PMID: 11179232
- Singh G, Rebbapragada I, Lykke-Andersen J. A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol 2008; 6:e111; PMID: 18447585
- Cosson B, Couturier A, Chabelskaya S, Kiktev D, Inge-Vechtomov S, Philippe M, Zhouravleva G. Poly(A)-binding protein acts in translation termination via eukaryotic release factor 3 interaction and does not influence [PSI(+)] propagation. Mol Cell Biol 2002; 22:3301 - 15; PMID: 11971964
- Hoshino S, Imai M, Kobayashi T, Uchida N, Katada T. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-Poly(A) tail of mRNA. Direct association of erf3/GSPT with polyadenylate-binding protein. J Biol Chem 1999; 274:16677 - 80; PMID: 10358005
- Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J 2008; 27:736 - 47; PMID: 18256688
- Behm-Ansmant I, Gatfield D, Rehwinkel J, Hilgers V, Izaurralde E. A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J 2007; 26:1591 - 601; PMID: 17318186
- Eberle AB, Stalder L, Mathys H, Orozco RZ, Mühlemann O. Posttranscriptional gene regulation by spatial rearrangement of the 3′ untranslated region. PLoS Biol 2008; 6:e92; PMID: 18447580
- Bühler M, Steiner S, Mohn F, Paillusson A, Mühlemann O. EJC-independent degradation of nonsense immunoglobulin-mu mRNA depends on 3′ UTR length. Nat Struct Mol Biol 2006; 13:462 - 4; PMID: 16622410
- Yepiskoposyan H, Aeschimann F, Nilsson D, Okoniewski M, Mühlemann O. Autoregulation of the nonsense-mediated mRNA decay pathway in human cells. RNA 2011; 17:2108 - 18; PMID: 22028362
- Johns L, Grimson A, Kuchma SL, Newman CL, Anderson P. Caenorhabditis elegans SMG-2 selectively marks mRNAs containing premature translation termination codons. Mol Cell Biol 2007; 27:5630 - 8; PMID: 17562857
- Kurosaki T, Maquat LE. Rules that govern UPF1 binding to mRNA 3′ UTRs. Proc Natl Acad Sci U S A 2013; 110:3357 - 62; PMID: 23404710
- Yamashita A, Izumi N, Kashima I, Ohnishi T, Saari B, Katsuhata Y, Muramatsu R, Morita T, Iwamatsu A, Hachiya T, et al. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev 2009; 23:1091 - 105; PMID: 19417104
- Metze S, Herzog VA, Ruepp MD, Muhlemann O. Comparison of EJC-enhanced and EJC-independent NMD in human cells reveals two partially redundant degradation pathways. RNA 2013; advanced online publication 20.8.2013.
- Kim VN, Kataoka N, Dreyfuss G. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon-exon junction complex. Science 2001; 293:1832 - 6; PMID: 11546873
- Le Hir H, Gatfield D, Izaurralde E, Moore MJ. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J 2001; 20:4987 - 97; PMID: 11532962
- Buchwald G, Ebert J, Basquin C, Sauliere J, Jayachandran U, Bono F, Le Hir H, Conti E. Insights into the recruitment of the NMD machinery from the crystal structure of a core EJC-UPF3b complex. Proc Natl Acad Sci U S A 2010; 107:10050 - 5; PMID: 20479275
- Melero R, Buchwald G, Castano R, Raabe M, Gil D, Lazaro M, Urlaub H, Conti E, Llorca O. The cryo-EM structure of the UPF-EJC complex shows UPF1 poised toward the RNA 3′ end. Nat Struct Mol Biol 2012; 19:498-505, S1-2.
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J 2000; 19:6860 - 9; PMID: 11118221
- Le Hir H, Moore MJ, Maquat LE. Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon-exon junctions. Genes Dev 2000; 14:1098 - 108; PMID: 10809668
- Barbosa I, Haque N, Fiorini F, Barrandon C, Tomasetto C, Blanchette M, Le Hir H. Human CWC22 escorts the helicase eIF4AIII to spliceosomes and promotes exon junction complex assembly. Nat Struct Mol Biol 2012; 19:983 - 90; PMID: 22961380
- Steckelberg AL, Boehm V, Gromadzka AM, Gehring NH. CWC22 connects pre-mRNA splicing and exon junction complex assembly. Cell Rep 2012; 2:454 - 61; PMID: 22959432
- Dostie J, Dreyfuss G. Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr Biol 2002; 12:1060 - 7; PMID: 12121612
- Gehring NH, Lamprinaki S, Kulozik AE, Hentze MW. Disassembly of exon junction complexes by PYM. Cell 2009; 137:536 - 48; PMID: 19410547
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr., Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 2010; 141:129 - 41; PMID: 20371350
- König J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol 2010; 17:909 - 15; PMID: 20601959
- Hurt JA, Robertson AD, Burge CB. Global analyses of UPF1 binding and function reveal expanded scope of nonsense-mediated mRNA decay. Genome Res 2013; 23:1636 - 50; PMID: 23766421
- Zünd D, Gruber AR, Zavolan M, Mühlemann O. Translation-dependent displacement of UPF1 from coding sequences causes its enrichment in 3′ UTRs. Nat Struct Mol Biol 2013; 20:936 - 43; PMID: 23832275
- Hogg JR, Goff SP. Upf1 senses 3’UTR length to potentiate mRNA decay. Cell 2010; 143:379 - 89; PMID: 21029861
- Sugimoto Y, König J, Hussain S, Zupan B, Curk T, Frye M, Ule J. Analysis of CLIP and iCLIP methods for nucleotide-resolution studies of protein-RNA interactions. Genome Biol 2012; 13:R67; PMID: 22863408
- Saulière J, Murigneux V, Wang Z, Marquenet E, Barbosa I, Le Tonquèze O, Audic Y, Paillard L, Roest Crollius H, Le Hir H. CLIP-seq of eIF4AIII reveals transcriptome-wide mapping of the human exon junction complex. Nat Struct Mol Biol 2012; 19:1124 - 31; PMID: 23085716
- Singh G, Kucukural A, Cenik C, Leszyk JD, Shaffer SA, Weng Z, Moore MJ. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell 2012; 151:750 - 64; PMID: 23084401
- Tani H, Imamachi N, Salam KA, Mizutani R, Ijiri K, Irie T, Yada T, Suzuki Y, Akimitsu N. Identification of hundreds of novel UPF1 target transcripts by direct determination of whole transcriptome stability. RNA Biol 2012; 9:1370 - 9; PMID: 23064114
- Hogg JR. This message was inspected by Upf1: 3’UTR length sensing in mRNA quality control. Cell Cycle 2011; 10:372 - 3; PMID: 21248469