Abstract
Research on transfer RNA (tRNA) has gone a long way since the existence of this essential adapter of the genetic code was first hypothesized five decades ago. With the new and fascinating discovering of connections between tRNAs and cellular pathways beyond genetic translation, the field of tRNA research has reached a new era. Here, we review some aspects of the emerging variety of tasks performed by full length tRNAs as well as their fragments generated by specific nuclease cleavage. Topics of special focus include the effect of differential expression of tRNAs in healthy tissues as well as their frequent deregulation observed in cancer cells. We also discuss the central role played by tRNAMet in cell metabolism, proliferation, and response to oxidative stress. Finally we review evidences suggesting that tRNAs are critical sources of short RNAs regulating an ever growing variety of cellular processes including translation initiation, control of genomic retroviral sequences, or RNA interference.
Introduction
In eukaryotes, tRNA genes are massively transcribed by RNA polymerase III. Millions of tRNAs molecules are synthesized per generation, which represent, at any time, at least 30% of all RNA in a cell.Citation1 tRNA transcription comes at an obvious and tremendous energetic cost and therefore needs to be tightly regulated in response to nutrient availability and cellular fitness.
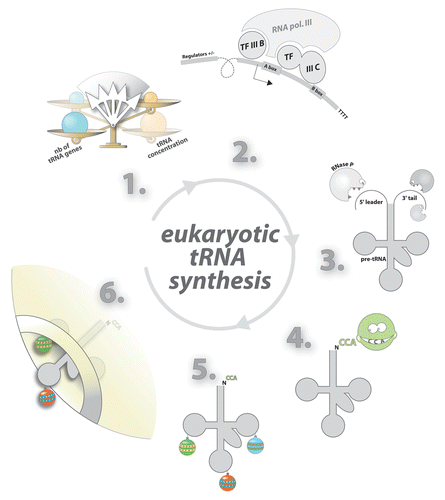
A portion of the RNA polymerase III promoter resides within each eukaryotic tRNA gene (). This internal promoter encompasses two distinct regions corresponding to nucleotides 8–19 (box A) and 52–62 (box B) of tRNA.Citation2 tRNA biogenesis involves the synthesis of an initial transcript containing a 5′ leader sequence, which is subsequently removed by the endonuclease RNase P.Citation3 Maturation of the 3′ end requires trimming the 3′ trailer and subsequent addition of CCA after the discriminator base N73Citation4 (). Only a small fraction of tRNA genes contain introns that are invariably found between nucleotides 37 and 38.Citation5 Splicing of tRNAs is essential and involves only a limited number of proteins as opposed to spliceosome-mediated mRNA splicing. Prior to their export from the nucleus to the cytoplasm, freshly processed tRNAs are extensively modified. On average 14 modifications are added post-transcriptionally in eukaryotic tRNAs.Citation6 The structure and the position of these modifications modulate tRNA activity to different extents. Modifications in or around the anticodon loop tune translation rate and impact cellular growth.Citation7 Modifications in the main body typically affect tRNA folding and stability.Citation8,Citation9
Figure 1. Cloverleaf folding of tRNA (tRNA). The standard cloverleaf structure (2D) and conventional numbering is shown. Conserved nucleotides are indicated (T and Ψ are modified residues: ribothymidine and pseudouridine; R are for purines and Y for pyrimidines). Watson-Crick base pairings and G•U pairs are shown; thin black lines indicate long-distance base pairings involved in tertiary folding (3D, upper right panel).
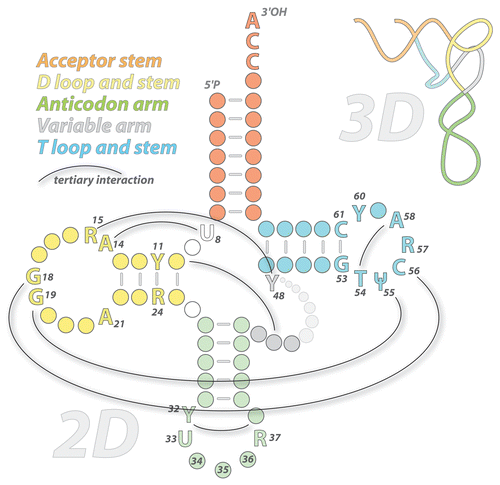
Figure 2. Synthesis, processing, and nucleo-cytoplasmic export of eukaryotic tRNA. In eukaryotes, tRNA concentrations fluctuate according to cell demand and are not proportional to the number tRNA genes in the genome (1). tRNA genes are transcribed by RNA polymerase (2) as long precursors (pre-tRNA). Additional sequences are then enzymatically removed (3). Functional tRNAs display a CCA sequence at their 3′-end. This triplet is added post-transcriptionally by the tRNA nucleotidyl transferase (CCase) (4). Base modifications are added at the final processing step (5). Export of tRNA through nuclear pores is facilitated by exportin-t and Ran-GTP (6).
Basal tRNA Expression in Human
Organization of tRNA genes in the human genome
tRNAs were long considered as archetypical house-keeping molecules. They were thought to serve an exclusive role in protein translation and supposedly lacked regulatory functions. Prior to the human genome-sequencing project, human tRNAs were expected to be no more diverse than those in unicellular organisms.
The human genome encodes around 500 tRNA genes, which represent five times the tRNA gene population in the bacteria E. coli. These genes are found throughout the genome and are present on all but the Y chromosome.Citation10 The mitochondrial DNA encodes an additional 22 tRNA genes. The corresponding tRNAs are grouped into 49 isoacceptor families and decode 21 amino acids. Interestingly, the largest cluster of tRNA genes resides in the gene cluster of the major histocompatibility complex also known as the leukocyte antigen complex. This co-localization suggests a potential coordination between canonical protein translation and immune system functions.Citation11
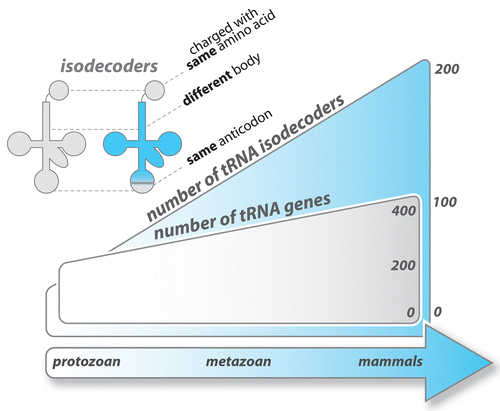
Another remarkable and unforeseen feature of tRNA that was revealed by genome sequencing is the proliferation of tRNA isodecoder genes. Isodecoders share identical anticodon sequence but display differences elsewhere in the tRNA body. The number and percentage of such tRNA genes follows remarkably the phylogenetic arrangement of living organisms. This fraction is < 10% in the budding yeast, 12–18% in fruit flies and worms and increases to 35–46% in chickens, dogs, rats, and mice.Citation10 In humans > 50% of tRNA genes are isodecoders (). Isodecoders have identical decoding capacity and therefore appear redundant at first glance. In reality, tRNA isodecoders display significant functional variations. An increasing body of evidence suggests that some isodecoders, particularly those with low affinity for the translation machinery, perform functions beyond protein synthesis and act as potent cell regulators.Citation12,Citation13
Figure 3. Number of tRNA genes and isodecoder genes across the phylogenetic spectrum. Eukaryotic genomes contain between 200 and 450 tRNA genes encoding 41 to 55 tRNA isoacceptors (tRNAs with different anticodons). The number of tRNA genes having the same anticodon but different sequences elsewhere in the tRNA body (tRNA isodecoders) varies significantly and is increasing across the phylogenetic spectrum. Sequence variations in tRNA isodecoders are concentrated in the internal promoter regions for RNA polymerase III.
Differential tRNA expression in healthy human tissues
Global tRNA expression is theoretically adjustable. tRNAs are transcribed by RNA polymerase III and its associated transcription factors, TFIIIB and TFIIIC. On one hand, intracellular abundance of these two essential factors is subjected to regulation as suggested by results from mRNA expression arrays.Citation14 Alternatively, TFIIIB and TFIIIC activities can be modulated at the post-translational level.Citation15
Regulation of the expression of individual isodecoders is also possible. Only a few nucleotides in the internal tRNA gene promoter are highly conserved because of their involvement in the tRNA tertiary structure. Seven nucleotides within box A and 6 nucleotides within box B are variable. These sequence differences could modulate the promoter strength by increasing or lowering the binding affinity for the aforementioned transcription factors.
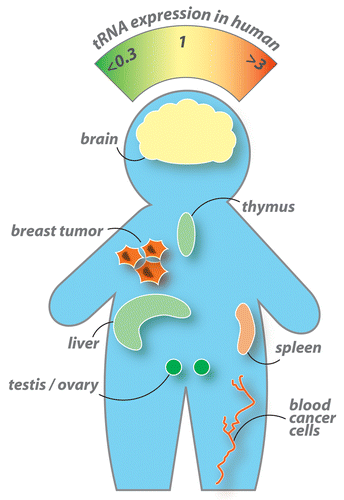
Comparative tRNA expression in brain, liver, vulva, testis, ovary, thymus, lymph node, and spleen tissue show that tRNA concentrations vary by as much as 10-fold among these eight organs.Citation11 The three immune tissues contain increased levels of tRNAs charged with hydrophobic amino acids and decreased levels of tRNAs for the charged group compared with brain tissue. In contrast, the two reproductive tissues contain decreased levels of tRNAs for the hydrophobic group and increased levels of tRNAs for the small side chain group. It appears that cellular tRNA concentrations naturally adjust to codon usage in order to maintain translation speed and limit ribosomal drop-off. Ribosomal proteins constitute by far the largest fraction of the proteome and therefore predominantly influence tRNA distribution.Citation16 Tissue-specific genes are sometimes expressed at a rate comparable to ribosomal components. Because these tissue-specific proteins are synthesized at such a high level they are directly responsible for the partial reprogramming of tRNA expression.
tRNAs are Overabundant in Cancer Cells
In multiple myeloma
In multiple myeloma (MM), malignant plasma cells overexpress cytosolic tRNAs. Levels of tRNAs derived from chromosomal-encoded genes are elevated by 2- to 4-fold, whereas the levels of tRNAs derived from mitochondrial-encoded genes remain unchangedCitation17 (). Myeloma cells are also characterized by the production and secretion of large amounts of monoclonal antibodies.
In breast cancer
tRNAs are invariably overexpressed in breast cancer cells and the corresponding expression patterns are remarkably conserved across the major subtypes of breast cancer. Cellular concentrations of nuclear- and mitochondrial-encoded tRNAs increase by up to 5-fold in breast cancer cell lines vs. non-cancer derived cell lines.Citation18 Interestingly, this difference is even more pronounced when malignant cells are compared with regular primary cells. Indeed, breast tumor cells express 10 times more nuclear- and mitochondrial-encoded tRNAs than the corresponding healthy breast tissue.
Functional consequence
Deregulation of RNA polymerase III and abnormal levels of pol III transcripts is common in a wide range of transformed cells. Cancer cells often overexpress TFIII factors, which subsequently lead to overexpression of tRNAs.Citation19,Citation20 In addition, polyploidy or genomic instability together with changes in chromatin structure drastically impact the expression of hundreds of genes in cancer relative to healthy cells. Chaotic and unpredictable tRNA expression in pathological cells would be an expected consequence. Surprisingly, transformed and cancerous cells preserve balanced tRNA pools that resemble the pools of healthy cells although they express considerably more tRNA molecules than regular cells.Citation16 In cancer cells, tRNA expression is regulated globally and simultaneously on the entire collection of tRNA genes.
Central Role for tRNAMet and MetRS
Artificial overexpression of initiator tRNAMet reprograms overall tRNA expression
Global regulation of tRNA expression in cancer cells enables simultaneous and coordinated transcription of all tRNAs. By definition, the same mechanism inhibits or keeps in check the overexpression of single tRNA species. Because tRNAs were long considered non-regulatory housekeeping genes, there are no well-established efficient methods for redistributing tRNA levels in mammalian cells. In addition, since tRNAs are so abundant, a mere 2-fold overexpression requires the biosynthesis and maturation of an additional 1,000,000 tRNAs per cell.Citation21 Artificial tRNA overexpression in cancer cells requires the generation of stable cell lines and only modest but nonetheless significant increase (1.4- to 2.2-fold) can be achieved. Interestingly, even low expression of additional initiator tRNAMet in non-tumorigenic cells increases cell metabolism and proliferation. A rush of initiator tRNA in the translation machinery may have two consequences. First, it may boost global mRNA translation and stimulate overall protein synthesis. Second, it may favor translation of mRNA encoding cell-cycle or anti-apoptotic proteins such as Myc or Cyclin D1 and drive the cell toward a more defined and pronounced pro-cancer state. Overexpression of initiator tRNAMet also globally increases tRNA levels, confirming the necessity to maintain a balanced pool of tRNAs in fast dividing cells. Between the two options of actively degrading excess of initiator tRNA and raising the rest of the tRNA pool up to the level of the initiator tRNA, cells choose the latter against all energetic considerations.
Misacylation of tRNA by methionyl-tRNA synthetase has been observed in bacteria, yeast, and mammals
tRNA-microarray analyses have demonstrated that methionyl-tRNA synthetases (MetRS) from Escherichia coli, Saccharomyces cerevisiae, and humans misacylate non-cognate tRNAs with methionine. These misacylated species are used in translation and lead to the synthesis of mutant proteins with potential new features or properties.
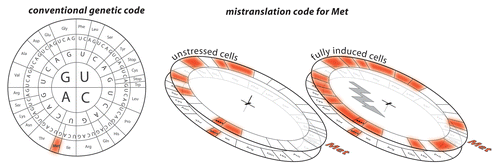
E. coli MetRS mismethionylates only two E. coli tRNA species in vitro, one tRNAThr and one tRNAArg. Strong and very specific interactions between residues of the enzyme and the anticodon stimulate or alternatively prevent the transfer of methionine onto the tRNA. Therefore, only a very limited set of non-methionine tRNAs display identity elements for mismethionylation.Citation22 On the contrary, S. cerevisiae MetRS mismethionylates a wide range of non-methionine tRNAs.Citation23 The magnitude of mismethionylation depends directly on growth conditions. At stationary phase, mismethionylation culminates and one in every ten methionine is transferred onto non-cognate tRNAs. The yeast MetRS is part of a unique three-protein complex that includes a general tRNA-binding protein, Arc1p and the glutamyl-tRNA synthetase. This complex equips yeast MetRS with strong and non-specific tRNA binding abilities explaining the extensive level of misacylated tRNAs in that species. In unstressed mammalian cells, approximately 1% of the methionylated tRNAs are non-cognate and mismethionylation is further increased by up to 10-fold when cells are exposed to various forms of oxidative stressCitation24(). Interestingly, mismethionylation was not observed for any yeast or human mitochondrial tRNA. Mismethionylation patterns of E. coli, yeast and mammals do not correspond to each other. Further studies may determine if mammalian mismethionylation is an evolutionary extension of bacterial and yeast pathways or represents the convergent evolution of a distinct mechanism.
Methionine misacylation protects cells against oxidative stress
Many cellular stressors trigger the release of reactive oxygen species (ROS). ROS damage cellular components in general and oxidize the highly reactive sulfur in methionine residues in particular. Proteins have evolved to minimize the impact of ROS by hiding functional residues such as histidine, tryptophan, cysteine or methionine behind protein motifs displaying decoy or non-essential methionine.Citation25 Mismethionylation and subsequent misincorporation of methionine residues in neo-synthesized proteins enhances the protective function of genetically encoded decoy methionine residues. This complementary pathway is induced rapidly and allows immediate extra-genetic incorporation of methionine in response to increased ROS levels. An obvious side effect of this mechanism is the apparent undesirable synthesis of catalytically inactive or misfolded proteins. These aberrant proteins constitute only a minor fraction of the mismethionylation-derived protein pools, as other proteins would contain extra methionine in positions with minimal disturbance to catalytic activity or structure. Rapid containment of ROS comes at a cost and these proteins are the result of a cellular compromise.
tRNA Fragments: The Challenge of Defining Nomenclature, Classification and Biological Functions
The first report describing the inhibitory role of a tRNA fragment isolated from human urinary bladder carcinoma was published in 1999.Citation26 Since this initial report, an eclectic collection of tRNA fragments have been found in bacteria, fungi, plants, and animals.Citation27-Citation37 All these fragments were indifferently described as tRNA halvesCitation30,Citation38 or tRNA-derived halves.Citation39 Acronyms such as tRFs(tRNA-derived RNA fragments),Citation36 tiRNAs (stress-induced small RNAs),Citation39,Citation40sitRNAs (stress-induced tRNAs),Citation33tsRNAs (tRNA-derived small RNAs)Citation37 and ubcRNAs (urinary bladdercarcinoma RNAs)Citation26 were used sometimes to refer to very similar tRNA sub-entities. As this collection of fragments grew exponentially, the definition of a unifying nomenclature became indispensable.Citation41 The two major classes of tRNA fragments are currently defined as tRNA halves and small tRNA fragments (tRFs).
tRNA halves: effector molecules of stress response
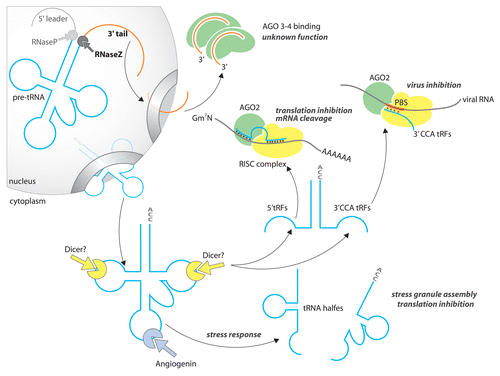
Angiogenin (ANG) and Rny1p generate tRNA halves of 30 to 35 nucleotides, in mammalsCitation39,Citation40 and yeastCitation42 respectively, by splitting mature tRNAs in or near their anticodon loop (). These cleavages are induced by starvation in Tetrahymena thermophila,Citation30 serum deprivation in Giardia lambia,Citation33 during development in Aspergillus fumigatusCitation43 or in response to oxidative stress in eukaryotes.Citation38,Citation42,Citation44 Most 5′ halves generated by ANG inhibit protein synthesis by inducing eIF2α-phosphorylation and promoting stress granule assembly.Citation40,Citation45,Citation46 Alternatively, some 5′ tRNA halves bind directly to eIF4G, eIF4A, and eIF4G/eIF4A complexes sequestering pivotal translation components away from mRNA.Citation45 This binding is driven by the presence of oligoguanine (TOG) motifs of four to five residues found at the 5′ end of tRNA halves.
Three subclasses of small tRNA fragments (tRFs): 5′, 3′ CCA, and 3′ U tRFs
The biosynthesis of 5′tRFs is complex and still cryptic in many aspects. Several nucleases are apparently responsible for the generation of fragments of different sizes (). It has been proposed that Dicer, the microRNA processing enzyme, may be involved in the production of 19 to 22-nucleotide tRFs.Citation34,Citation37 Interestingly, Dicer knockout still supports the production of 10 to 15 nucleotides tRFsCitation35 suggesting the existence of a Dicer-independent pathway for smaller fragments. Finally, it has been proposed that angiogenin, which cuts primarily within the anticodon loop, could potentially also target both the D-loop and the TψU-loop in order to generate ~20-nt long 5′ and 3′ tRFs.Citation35 Because 5′ tRFs are produced by Dicer and found associated with AGO2Citation34 one would expect them to have miRNA-like activities. Surprisingly, 5′tRFs derived from tRNAGln inhibit the translation of reporter genes in vivo and in vitro although they intrinsically have little to no ability to base pair with their primary targets.Citation42 An unexpected and recent report has shown that a 5′tRF derived from tRNAVal downregulates protein synthesis in the archaeon Haloferax volcanii by interfering with the small ribosomal subunit.Citation47
3′ CCA tRFs are emerging cellular regulators with tremendous biomedical potential. Retroviruses like the Human immunodeficiency virus (HIV-1) highjack cellular tRNALys, tRNAPro, or tRNATrp in order to replicate their genome. In other words, these tRNAs bind to retroviral primer binding site (PBS) and serve as primers to initiate reverse transcription.Citation39,Citation40 Interestingly, 3′ CCA tRFs originating from human tRNALys maintain their PBS binding ability.Citation25 It was initially believed that these fragments could be used as antiviral countermeasures by competing with tRNALys. A report published in 2009 revealed the true extent of 3′CCA tRFs’ regulatory functions. Viral PBS base pairing with these fragments becomes excellent substrate for DicerCitation26 (). The cleavage of PBS gives rise to another short RNA named PBSnc RNA, which inhibits HIV replication most likely through RNAi pathways. A significant fraction of the human genome is made of relics of retroviral sequences. Many identified 3′ tRFs are highly complementary to endogenous retroviral PBS. tRNA fragments constitute a potential reservoir of molecules able to keep the expression of viral sequences in check.Citation10
3′U tRFs are derived from the 3′ trailer of tRNA precursors (). These fragments typically display five to six consecutive uridine residues at their 3′ end, which correspond to the canonical RNA polymerase III stop signal. 3′U tRFs are found exclusively in the cytoplasm suggesting instant export from the nucleus to the cytoplasm or alternative pre-tRNA processing by a cytosolic form of RNase Z.Citation37,Citation50 3′U tRFs are essential and potent regulators of cellular proliferation, they are consistently overexpressed in cancer cell lines. RNaseZ, an enzyme essential for their biogenesis, is also known as the prostate cancer susceptibility gene ELAC2.Citation36 3′U tRFs bind preferentially the non-splicing AGO3 and 4. The cellular impact of these fragments is therefore RNAi independent and likely to be indirect. First, the saturation of AGO3 and 4 could potentiate the silencing activity of miRNA by favoring their binding to effector AGO1 and 2. Second, AGO3 and 4 are involved in various cellular pathways and their binding to 3′Υ tRFs could modulate their activity. For example, AGO4 stimulates spermatogenesis and regulates cell cycle.Citation51 AGO3 participates in Alu RNA guided mRNA decay during stem cell proliferation.Citation52
Conclusion
Five decades of intensive research has revealed the function of tRNAs in protein biosynthesis at the atomic, molecular and cellular level. The recent standardization of high throughput approaches opens whole new perspectives by boosting the discovery of new tRNA functions and allowing the scientific community to appreciate the extent of their ramifications. New functions could not have been envisioned due to their intricacy and apparent functional distance to the translation machinery. tRNAs have tremendous potential for cellular regulation as a full-length molecule but also when shattered into pieces. How an ancient housekeeping RNA evolved to become such a potent, versatile, and specific cellular regulator still remains elusive. However some obvious tRNA features have most certainly facilitated or even driven this transition. First and foremost, tRNAs are extremely abundant and consequently highly redundant. A significant number of tRNA molecules can be rechanneled, away from the translation machinery, without perturbing the dynamic of protein synthesis. Second, tRNA sequences are highly malleable. Myriads of compensatory mutations allow the shuffling of nucleotides without affecting functional folding. Finally, this ubiquitous and primordial molecule coevolved with literally every living organism. This early coexistence gave tRNAs the unique opportunity to interact with a wide variety of cellular mechanisms and grow deep ramifications within modern pathways. The cellular network of molecular interactions constitutes in many ways the utmost jigsaw. Tremendous progress has been made toward solving this puzzle and many parts have been assembled independently of each other. TRNAs could be one of the puzzle pieces needed to kilt these parts together.
| Abbreviations: | ||
| tRNA | = | transfer RNA molecule |
| RNase | = | ribonuclease |
| ROS | = | reactive oxygen species |
| MetRS | = | methionyl-tRNA synthetases |
| tRFs | = | tRNA-derived RNA fragments |
| PBS | = | primer binding site |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The College of Charleston and Centre National de la Recherche Scientifique (ANR11SVSE8–025–01) sponsor the authors’ current research activities. We thank Jomel Jacinto and Joseph Karam from the College of Charleston for comments on this manuscript.
References
- Waldron C, Lacroute F. Effect of growth rate on the amounts of ribosomal and transfer ribonucleic acids in yeast. J Bacteriol 1975; 122:855 - 65; PMID: 1097403
- Galli G, Hofstetter H, Birnstiel ML. Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature 1981; 294:626 - 31; http://dx.doi.org/10.1038/294626a0; PMID: 7312050
- Walker SC, Engelke DR. Ribonuclease P: the evolution of an ancient RNA enzyme. Crit Rev Biochem Mol Biol 2006; 41:77 - 102; http://dx.doi.org/10.1080/10409230600602634; PMID: 16595295
- Dubrovsky EB, Dubrovskaya VA, Levinger L, Schiffer S, Marchfelder A. Drosophila RNase Z processes mitochondrial and nuclear pre-tRNA 3′ ends in vivo. Nucleic Acids Res 2004; 32:255 - 62; http://dx.doi.org/10.1093/nar/gkh182; PMID: 14715923
- Marck C, Grosjean H. tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA 2002; 8:1189 - 232; http://dx.doi.org/10.1017/S1355838202022021; PMID: 12403461
- Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, et al. MODOMICS: a database of RNA modification pathways--2013 update. Nucleic Acids Res 2013; 41:D262 - 7; http://dx.doi.org/10.1093/nar/gks1007; PMID: 23118484
- El Yacoubi B, Hatin I, Deutsch C, Kahveci T, Rousset JP, Iwata-Reuyl D, Murzin AG, de Crécy-Lagard V. A role for the universal Kae1/Qri7/YgjD (COG0533) family in tRNA modification. EMBO J 2011; 30:882 - 93; http://dx.doi.org/10.1038/emboj.2010.363; PMID: 21285948
- Sengupta R, Vainauskas S, Yarian C, Sochacka E, Malkiewicz A, Guenther RH, Koshlap KM, Agris PF. Modified constructs of the tRNA TPsiC domain to probe substrate conformational requirements of m(1)A(58) and m(5)U(54) tRNA methyltransferases. Nucleic Acids Res 2000; 28:1374 - 80; http://dx.doi.org/10.1093/nar/28.6.1374; PMID: 10684932
- Helm M, Giegé R, Florentz C. A Watson-Crick base-pair-disrupting methyl group (m1A9) is sufficient for cloverleaf folding of human mitochondrial tRNALys. Biochemistry 1999; 38:13338 - 46; http://dx.doi.org/10.1021/bi991061g; PMID: 10529209
- Goodenbour JM, Pan T. Diversity of tRNA genes in eukaryotes. Nucleic Acids Res 2006; 34:6137 - 46; http://dx.doi.org/10.1093/nar/gkl725; PMID: 17088292
- Dittmar KA, Goodenbour JM, Pan T. Tissue-specific differences in human transfer RNA expression. PLoS Genet 2006; 2:e221; http://dx.doi.org/10.1371/journal.pgen.0020221; PMID: 17194224
- Geslain R, Pan T. Functional analysis of human tRNA isodecoders. J Mol Biol 2010; 396:821 - 31; http://dx.doi.org/10.1016/j.jmb.2009.12.018; PMID: 20026070
- Rudinger-Thirion J, Lescure A, Paulus C, Frugier M. Misfolded human tRNA isodecoder binds and neutralizes a 3′ UTR-embedded Alu element. Proc Natl Acad Sci U S A 2011; 108:E794 - 802; http://dx.doi.org/10.1073/pnas.1103698108; PMID: 21896722
- Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, et al. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci U S A 2002; 99:4465 - 70; http://dx.doi.org/10.1073/pnas.012025199; PMID: 11904358
- Conesa C, Swanson RN, Schultz P, Oudet P, Sentenac A. On the subunit composition, stoichiometry, and phosphorylation of the yeast transcription factor TFIIIC/tau. J Biol Chem 1993; 268:18047 - 52; PMID: 7688737
- Mahlab S, Tuller T, Linial M. Conservation of the relative tRNA composition in healthy and cancerous tissues. RNA 2012; 18:640 - 52; http://dx.doi.org/10.1261/rna.030775.111; PMID: 22357911
- Zhou Y, Goodenbour JM, Godley LA, Wickrema A, Pan T. High levels of tRNA abundance and alteration of tRNA charging by bortezomib in multiple myeloma. Biochem Biophys Res Commun 2009; 385:160 - 4; http://dx.doi.org/10.1016/j.bbrc.2009.05.031; PMID: 19450555
- Pavon-Eternod M, Gomes S, Geslain R, Dai Q, Rosner MR, Pan T. tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res 2009; 37:7268 - 80; http://dx.doi.org/10.1093/nar/gkp787; PMID: 19783824
- Winter AG, Sourvinos G, Allison SJ, Tosh K, Scott PH, Spandidos DA, White RJ. RNA polymerase III transcription factor TFIIIC2 is overexpressed in ovarian tumors. Proc Natl Acad Sci U S A 2000; 97:12619 - 24; http://dx.doi.org/10.1073/pnas.230224097; PMID: 11058163
- Marshall L, White RJ. Non-coding RNA production by RNA polymerase III is implicated in cancer. Nat Rev Cancer 2008; 8:911 - 4; http://dx.doi.org/10.1038/nrc2539; PMID: 18987635
- Pavon-Eternod M, Gomes S, Rosner MR, Pan T. Overexpression of initiator methionine tRNA leads to global reprogramming of tRNA expression and increased proliferation in human epithelial cells. RNA 2013; 19:461 - 6; http://dx.doi.org/10.1261/rna.037507.112; PMID: 23431330
- Jones TE, Alexander RW, Pan T. Misacylation of specific nonmethionyl tRNAs by a bacterial methionyl-tRNA synthetase. Proc Natl Acad Sci U S A 2011; 108:6933 - 8; http://dx.doi.org/10.1073/pnas.1019033108; PMID: 21482813
- Wiltrout E, Goodenbour JM, Fréchin M, Pan T. Misacylation of tRNA with methionine in Saccharomyces cerevisiae. Nucleic Acids Res 2012; 40:10494 - 506; http://dx.doi.org/10.1093/nar/gks805; PMID: 22941646
- Netzer N, Goodenbour JM, David A, Dittmar KA, Jones RB, Schneider JR, Boone D, Eves EM, Rosner MR, Gibbs JS, et al. Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature 2009; 462:522 - 6; http://dx.doi.org/10.1038/nature08576; PMID: 19940929
- Pan T. Adaptive translation as a mechanism of stress response and adaptation. Annu Rev Genet 2013; 47:121 - 37; http://dx.doi.org/10.1146/annurev-genet-111212-133522; PMID: 23988117
- Zhao H, Bojanowski K, Ingber DE, Panigrahy D, Pepper MS, Montesano R, Shing Y. New role for tRNA and its fragment purified from human urinary bladder carcinoma conditioned medium: inhibition of endothelial cell growth. J Cell Biochem 1999; 76:109 - 17; http://dx.doi.org/10.1002/(SICI)1097-4644(20000101)76:1<109::AID-JCB11>3.0.CO;2-K; PMID: 10581005
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev 2008; 22:2773 - 85; http://dx.doi.org/10.1101/gad.1705308; PMID: 18923076
- Calabrese JM, Seila AC, Yeo GW, Sharp PA. RNA sequence analysis defines Dicer’s role in mouse embryonic stem cells. Proc Natl Acad Sci U S A 2007; 104:18097 - 102; http://dx.doi.org/10.1073/pnas.0709193104; PMID: 17989215
- Kawaji H, Nakamura M, Takahashi Y, Sandelin A, Katayama S, Fukuda S, Daub CO, Kai C, Kawai J, Yasuda J, et al. Hidden layers of human small RNAs. BMC Genomics 2008; 9:157; http://dx.doi.org/10.1186/1471-2164-9-157; PMID: 18402656
- Lee SR, Collins K. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J Biol Chem 2005; 280:42744 - 9; http://dx.doi.org/10.1074/jbc.M510356200; PMID: 16272149
- Zhang S, Sun L, Kragler F. The phloem-delivered RNA pool contains small noncoding RNAs and interferes with translation. Plant Physiol 2009; 150:378 - 87; http://dx.doi.org/10.1104/pp.108.134767; PMID: 19261735
- Haiser HJ, Karginov FV, Hannon GJ, Elliot MA. Developmentally regulated cleavage of tRNAs in the bacterium Streptomyces coelicolor. Nucleic Acids Res 2008; 36:732 - 41; http://dx.doi.org/10.1093/nar/gkm1096; PMID: 18084030
- Li Y, Luo J, Zhou H, Liao JY, Ma LM, Chen YQ, Qu LH. Stress-induced tRNA-derived RNAs: a novel class of small RNAs in the primitive eukaryote Giardia lamblia. Nucleic Acids Res 2008; 36:6048 - 55; http://dx.doi.org/10.1093/nar/gkn596; PMID: 18820301
- Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, Green PJ, Barton GJ, Hutvagner G. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 2009; 15:2147 - 60; http://dx.doi.org/10.1261/rna.1738409; PMID: 19850906
- Li Z, Ender C, Meister G, Moore PS, Chang Y, John B. Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs. Nucleic Acids Res 2012; 40:6787 - 99; http://dx.doi.org/10.1093/nar/gks307; PMID: 22492706
- Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 2010; 16:673 - 95; http://dx.doi.org/10.1261/rna.2000810; PMID: 20181738
- Fu H, Feng J, Liu Q, Sun F, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett 2009; 583:437 - 42; http://dx.doi.org/10.1016/j.febslet.2008.12.043; PMID: 19114040
- Garcia-Silva MR, Frugier M, Tosar JP, Correa-Dominguez A, Ronalte-Alves L, Parodi-Talice A, Rovira C, Robello C, Goldenberg S, Cayota A. A population of tRNA-derived small RNAs is actively produced in Trypanosoma cruzi and recruited to specific cytoplasmic granules. Mol Biochem Parasitol 2010; 171:64 - 73; http://dx.doi.org/10.1016/j.molbiopara.2010.02.003; PMID: 20156490
- Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol 2009; 185:35 - 42; http://dx.doi.org/10.1083/jcb.200811106; PMID: 19332886
- Emara MM, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N, Hu GF, Anderson P. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem 2010; 285:10959 - 68; http://dx.doi.org/10.1074/jbc.M109.077560; PMID: 20129916
- Sobala A, Hutvagner G. Small RNAs derived from the 5′ end of tRNA can inhibit protein translation in human cells. RNA Biol 2013; 10:553 - 63; http://dx.doi.org/10.4161/rna.24285; PMID: 23563448
- Thompson DM, Parker R. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J Cell Biol 2009; 185:43 - 50; http://dx.doi.org/10.1083/jcb.200811119; PMID: 19332891
- Jöchl C, Rederstorff M, Hertel J, Stadler PF, Hofacker IL, Schrettl M, Haas H, Hüttenhofer A. Small ncRNA transcriptome analysis from Aspergillus fumigatus suggests a novel mechanism for regulation of protein synthesis. Nucleic Acids Res 2008; 36:2677 - 89; http://dx.doi.org/10.1093/nar/gkn123; PMID: 18346967
- Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 2008; 14:2095 - 103; http://dx.doi.org/10.1261/rna.1232808; PMID: 18719243
- Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell 2011; 43:613 - 23; http://dx.doi.org/10.1016/j.molcel.2011.06.022; PMID: 21855800
- Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell 2009; 36:932 - 41; http://dx.doi.org/10.1016/j.molcel.2009.11.020; PMID: 20064460
- Gebetsberger J, Zywicki M, Kunzi A, Polacek N. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea 2012; 2012:260909.
- Goodchild J, Agrawal S, Civeira MP, Sarin PS, Sun D, Zamecnik PC. Inhibition of human immunodeficiency virus replication by antisense oligodeoxynucleotides. Proc Natl Acad Sci U S A 1988; 85:5507 - 11; http://dx.doi.org/10.1073/pnas.85.15.5507; PMID: 3041414
- Yeung ML, Bennasser Y, Watashi K, Le SY, Houzet L, Jeang KT. Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res 2009; 37:6575 - 86; http://dx.doi.org/10.1093/nar/gkp707; PMID: 19729508
- Elbarbary RA, Takaku H, Uchiumi N, Tamiya H, Abe M, Takahashi M, Nishida H, Nashimoto M. Modulation of gene expression by human cytosolic tRNase Z(L) through 5′-half-tRNA. PLoS One 2009; 4:e5908; http://dx.doi.org/10.1371/journal.pone.0005908; PMID: 19526060
- Modzelewski AJ, Holmes RJ, Hilz S, Grimson A, Cohen PE. AGO4 regulates entry into meiosis and influences silencing of sex chromosomes in the male mouse germline. Dev Cell 2012; 23:251 - 64; http://dx.doi.org/10.1016/j.devcel.2012.07.003; PMID: 22863743
- Hu Q, Tanasa B, Trabucchi M, Li W, Zhang J, Ohgi KA, Rose DW, Glass CK, Rosenfeld MG. DICER- and AGO3-dependent generation of retinoic acid-induced DR2 Alu RNAs regulates human stem cell proliferation. Nat Struct Mol Biol 2012; 19:1168 - 75; http://dx.doi.org/10.1038/nsmb.2400; PMID: 23064648