Abstract
The DEAD box RNA helicase DHH1 acts as a general repressor of translation and activator of decapping but can also act specifically on individual mRNAs. In trypanosomes, DHH1 overexpression or expression of a dhh1 ATPase mutant, dhh1 DEAD:DQAD, resulted in increased or decreased stability of a small group of mRNAs, mainly encoding developmentally regulated genes. Here, four of the mRNAs affected by dhh1 DEAD:DQAD expression have been analyzed to identify cis-elements involved in dhh1 DEAD:DQAD action. For three mRNAs, the 3′ UTR mediated the change in mRNA level and, in one case, both the 5′ and the 3′ UTR contributed. No responsive elements were detected in the protein coding sequences. One mRNA stabilized by dhh1 DEAD:DQAD expression was analyzed in more detail: deletion or mutation of an AU-rich element in the 3′ UTR resulted in mRNA stabilization in the absence of dhh1 DEAD:DQAD and completely abolished the response to dhh1 DEAD:DQAD. While AU-rich instability elements have been previously shown to mediate mRNA decrease or translational exit by recruitment of DHH1, this is, to our knowledge, the first report of an AU-rich instability element that is responsible for a DHH1 mediated increase in mRNA stability. We suggest a novel model for the selective action of dhh1 on individual mRNAs that is based on the change in the turnover rate of stabilizing or destabilizing RNA binding proteins.
Introduction
DEAD box RNA helicases bind to and translocate along RNA molecules by a ratchet-like mechanism. The effect of translocation can be to unwind short RNA duplexes and/or disrupt RNP complexes through displacement of RNA-binding proteins. Remodelling of RNP complexes in response to intrinsic or extrinsic signals almost certainly requires RNA helicase activity. The structure of DEAD box RNA helicases contains two lobes, each consisting of a RecA domain. RNA helicase activity uses cycles of ATP binding and hydrolysis.Citation1 RNA and ATP bind to the helicase cooperatively and force the two RecA like domains into a closed confirmation.Citation1,Citation2 RNA binding also stimulates ATP hydrolysis, which is not needed for the helicase activity of all enzymes tested to date, but rather required for the release from RNA.Citation3-Citation5 Some RNA helicases target specific mRNAs, others act as general chaperones in various stages of mRNA metabolism.
The RNA helicase DHH1, also known as Rck and p54, is conserved in all eukaryotesCitation6 and binds to RNA independently of sequenceCitation5,Citation7 with some preference for the 5′ UTR of mRNAs.Citation7 Like other DEAD box helicases, DHH1 has an unwinding activityCitation5,Citation8 which requires ATP binding, but not hydrolysis.Citation5 DHH1 has an RNA-stimulated ATPase activity in vitro but, at least for the yeast DHH1, this activity is unusually low and ATP does not enhance RNA binding.Citation5,Citation9 Invertebrate DHH1 homologs consist of only the core helicase motifs, yeast homologs have C-terminal extensions whereas vertebrate helicases have N-terminal extensions.Citation10 These extensions may serve organism specific functions, such as the binding to a specific RNA binding protein.Citation11
DHH1/Rck/p54 plays an important function in regulating the fate of mRNAs, which is reflected by the nature of its direct binding partners: proteins involved in mRNA decay (DCP1/DCP2, the LSM2–8 complex, EDC3, PAT1, CAF1) or translation (eEF1A).Citation12-Citation15 The exact function of the helicase is still a matter of debate. Initially, yeast DHH1 was found to act as a general activator of mRNA decapping.Citation16,Citation17 However, under conditions of no or low decapping activity DHH1 acts exclusively as a repressor of translation.Citation18-Citation21 Recent data from yeast suggest that DHH1 slows down ribosome movement, which could influence both decapping as well as translation.Citation21
In addition to its general functions, DHH1 can directly bind to several RNA binding proteins and is selectively recruited to individual mRNAs mediating translational exit and/or mRNA decay. One example is the cytoplasmic polyadenylation element binding protein (CPEB) that regulates the translation of some mRNAs during development. CPEB is found in the same complex as the Xenopus DHH1 ortholog, Xp54,Citation18,Citation22 and marks target mRNAs for DHH1-mediated translational repression. In yeast, DHH1 binds RBP1; both DHH1 and RBP1 mediate the instability of the target mRNA POR1, encoding porin.Citation11,Citation23 AU-rich elements (AREs) mediate mRNA instability via ARE binding proteins. The AU-rich element binding protein tristetraprolin (TTP) directly binds to and recruits DHH1 to its target mRNAs resulting in mRNA destabilization.Citation24 In a similar manner, iron-responsive mRNAs are destabilized by the ARE-binding proteins CTH1 and CTH2 in a manner dependent on an interaction between DHH1 and CTH2.Citation25
Trypanosomes are single cell flagellates responsible for Human African Trypanosomiasis and a range of livestock diseases. Several transitions between different developmental forms occur during the life-cycle in the mammalian and insect host, each transition is accompanied by significant changes in gene expression.Citation26-Citation29 The two forms that can be cultured in the lab are the blood stream forms (BSF), and the procyclic forms (PCF), representing proliferative stages from the mammalian and tsetse fly hosts, respectively. Maturation of mRNAs in trypanosomes is different to most other eukaryotes; all mRNAs are transcribed from polycistrons at roughly the same rate and processed by trans-splicing to add a capped short exon to the 5′ end, the 3′ end arises by endonuclease cleavage and polyadenylation.Citation30-Citation33 The constitutive transcription means that selective regulation of gene expression must occur post-transcriptionally with the one exception of a limited number of genes that are transcribed by RNA polymerase I.Citation34 Control of gene expression is believed to occur mainly by regulating translation and mRNA decay.Citation35-Citation38 Strikingly, although a decapping activity was reported39, trypanosomes lack orthologs to all proteins of the decapping complex, DCP1, DCP2, EDC3 and PAT1. Orthologs may not be identified in standard database searches due to the large evolutionary distance between trypanosomes and yeast/mammals, but the absence of all genes from the decapping complex rather indicates the absence of the entire complex.
Trypanosoma brucei DHH1 has 69% identity to yeast DHH1 but has no N or C-terminal extension.Citation40 DHH1 function in trypanosomes was studied by the expression of an ATPase mutant dhh1 DEAD:DQAD (dhh1 E182Q). In yeast, this mutant is impaired in ATP hydrolysis, but not in ATP bindingCitation9 and for the recombinant human DHH1 homolog Rck/p54 it was shown that ATP binding, but not ATP hydrolysis is needed for the unwinding activity of the helicase.Citation5 Instead, ATP hydrolysis may be required for the release of DHH1/Rck/p54 from mRNAs, as was shown for other DEAD box RNA helicases.Citation3,Citation4 Taken together, the data suggest that trypanosome dhh1 DEAD:DQAD is an active RNA helicase that may have an extended processivity in comparison to wild type DHH1.
Inducible expression of the ATPase mutant dhh1 DEAD:DQAD in a DHH1 wild type background resulted in a fast-onset growth arrest, global repression of translation and an increase in P-bodies, with only a minor reduction in total mRNA levels.Citation40 This suggests that T. brucei DHH1 mainly functions as a translational repressor, rather than as a promoter of the 5′-3′ mRNA decay pathway. However, DHH1 was originally identified in an RNAi screen as a factor required for the instability of a developmentally regulated mRNA.Citation40 In fact, transcriptome analysis revealed that dhh1 DEAD:DQAD expression had profound effects on a selected set of mRNAs: 26 mRNAs were > 2.5-fold upregulated and 31 were > 2-fold downregulated, in some cases regulation was > 10-fold. The increase in mRNA levels, for the three mRNAs tested, correlated with an increase in mRNA half-life and therefore resulted from an increase in mRNA stability.Citation40
At least one of the upregulated mRNAs, ISG75, remained in polysomes, apparently escaping the general translational repression. These data suggest that DHH1 may stabilize selected mRNAs by preventing their exit from polysomes. In agreement with the proposal that the dhh1 DEAD:DQAD mutant has an increased processivity on RNA, the phenotype obtained by dhh1 DEAD:DQAD expression can be copied in a weaker form by overexpression of the DHH1 wild type protein. The dhh1-sensitive set of mRNAs was significantly enriched for developmentally regulated mRNAs. The experiment was performed in the procyclic developmental form and mRNAs normally unstable in the procyclic life-cycle stage were stabilized. In contrast, developmentally regulated mRNAs selectively stable in the procyclic stage were destabilized. Together, dhh1 DEAD:DQAD expression resulted in a reversal of developmental regulation.
This study aimed to gain further insight into the mechanism underlying the dhh1 DEAD:DQAD mediated stabilization and destabilization of selected mRNAs in trypanosomes. Four previously identified dhh1 target mRNAs were analyzed for cis-acting elements necessary for the response to dhh1 DEAD:DQAD expression. In each case, the open reading frame was not required, whereas the 3′ UTRs were necessary and in one case both the 5′ and 3′ UTR mediated the dhh1 induced changes to a reporter mRNA. For one of the mRNAs stabilized by dhh1 DEAD:DQAD, the cis-acting element could be narrowed down to a short AU-rich element in the 3′UTR that acted as an instability element. However, this element did not mediate the developmental regulation of the mRNA. This is the first report of an AU-rich instability element that is responsible for dhh1 induced mRNA stabilization, rather than destabilization. We propose that an increase in dhh1 helicase activity will increase the rate of turnover of destabilizing factors, this way selectively stabilizing mRNAs that are otherwise unstable.
Results
Expression of dhh1 DEAD:DQAD altered the half life and thus steady-state levels of a set of mostly developmentally regulated mRNAs in trypanosomes.Citation40 To investigate the mechanism, four of these mRNAs were chosen for further analysis. Tb927.3.3730 (a putative ABC transporter) and Tb927.11.12100 (RNA-binding protein 5, RBP5) were normally unstable in the procyclic stage but were stabilized by the expression of dhh1 DEAD:DQAD. The other two, Tb927.10.15410 (glycosomal malate dehydrogenase, gMDH) and Tb927.6.2790 (a putative l-threonine 3-dehydrogenase), were destabilized by the expression of dhh1 DEAD:DQAD. The mRNAs are shown schematically in ; UTR lengths were taken from RNA sequencing data.Citation41
Figure 1. Sizes of the DHH1 target mRNAs used in this work. The UTR lengths are based on the RNA sequencing data ofCitation41 . When alternative splice acceptor sites or polyadenylation sites were identified, the length of the most abundant is shown in bold.

The open reading frames are not involved in DHH1-mediated changes in mRNA stability
Trypanosomes are diploid and any role of the open reading frame (ORF) in the DHH1-mediated change in mRNA levels was tested by replacing the ORF of one allele of the targeted gene with a puromycin acetyl transferase (PAC) ORF. The mRNAs from the resultant transgene contained the 5′ UTR from the endogenous gene followed by the PAC ORF followed by the 3′UTR from the endogenous gene. The transgene was introduced into the previously described cell line engineered for inducible expression of dhh1 DEAD:DQADCitation40 (). Puromycin resistant cell lines were obtained for three genes but not for Tb927.3.3730, this gene was excluded from this first analysis after several attempts to obtain the cell line failed.
Figure 2. ORF sequences are not necessary for the response to dhh1 DEAD:DQAD induction. (A) The locus of dhh1 DEAD:DQAD responsive genes was modified so that one allele remained wild type and the second allele was replaced with a puromycin acetyl transferase (PAC) ORF. A probe derived from the common 3′UTR was used to determine changes in steady-state mRNA levels on northern blots. (B) northern blots of total RNA from cell lines before and after dhh1 DEAD:DQAD induction (0 or 24 h TET) containing either an unmodified locus (WT/WT) or a modified locus (WT/PAC). Three independent clones of each cell lines were analyzed for each target gene. One representative northern blot is shown with rRNA used as a loading control. The average change in mRNA levels for both the transgene and the wild type gene was calculated from the three independent clones; error bars indicate standard errors. The expected sizes of the wild type and transgenic mRNAs are summarized in Supplemental Figure S1.
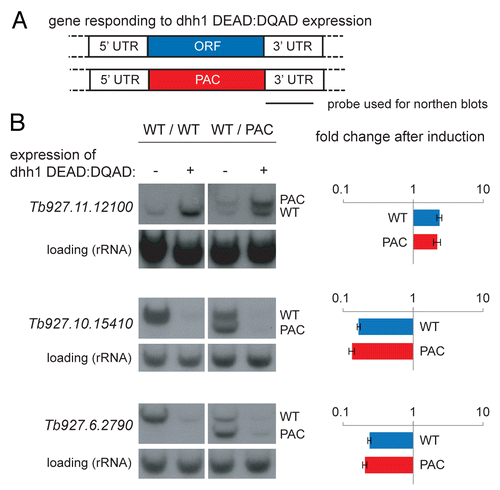
RNA was isolated from each of the three cell lines before and after induction of dhh1 DEAD:DQAD expression. Both the wild type mRNA as well as the transgenic mRNA with the PAC ORF were detected on the same northern blot by probing for the 3′ UTR; in each case, the size difference between the two mRNAs was sufficiently large to obtain separated signals () (Supplemental Fig. S1). RNA from the parental cell line, before and after induction of dhh1 DEAD:DQAD served as additional control. There was no significant difference in the dhh1 induced increase or decrease in mRNA levels between the mRNAs with the wild type open reading frames and the mRNAs containing the PAC open reading frames (). At least for the three selected mRNAs, the open reading frame is not involved in the response to dhh1 DEAD:DQAD expression.
Effect of the UTRs
The possible role of the UTRs in the response to dhh1 DEAD:DQAD was examined by attaching one or both UTRs to a reporter gene, the endogenous eIF4E3 gene. Using a cell line containing a tetracycline-inducible dhh1 DEAD:DQAD transgene,Citation40 one allele of the reporter was modified to replace the 5′ and/or 3′ UTR with the UTRs from each of the four target genes ( and Supplemental Fig. S2). RNA was prepared from cells before and after dhh1 DEAD:DQAD induction and both the mRNA arising from the wild type and the modified reporter alleles were detected on the same northern blot by a probe antisense to the open reading frame of the reporter gene (). In addition, northern blots with the same RNA samples were also probed for the target genes, using a probe antisense to the ORF probe for each gene ().
Figure 3. UTRs confer the response to dhh1 DEAD:DQAD on a reporter. (A) The locus of the reporter was modified so that one allele was wild type and the second contained either the 5′UTR or the 3′UTR of the dhh1 DEAD:DQAD responsive mRNA. A probe derived from the reporter ORF was used to measure steady-state levels on northern blots. See Supplemental Figure S2 for more details. (B) northern blots of total RNA before and after dhh1 DEAD:DQAD induction from cell lines with a wild type locus (R/R) or carrying one allele with the 5′UTR modified (R/5′UTR) or one allele with a modified 3′UTR (R/3′UTR) before and after induction of dhh1 DEAD:DQAD (0 or 24 h TET). The reporter gene mRNA as well as the mRNAs resulting from the modified loci were detected on the same blot by a probe detecting the reporter gene open reading frame. Northern blots with the same RNA were also probed for the respective DHH1 target genes. rRNA served as a loading control. One representative northern blot of three independent clones of each cell lines is shown as well as the quantification of the average change in mRNA level at dhh1 DEAD:DQAD expression for all transcripts (C). The error bars indicate standard errors. The expected sizes of the wild type and transgenic mRNAs are summarized in Supplemental Figure S1.
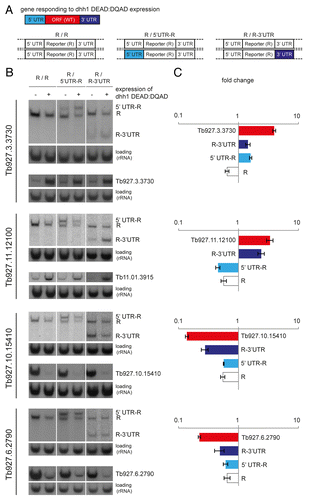
The effect of dhh1 DEAD:DQAD expression on the wild type reporter mRNA was a decrease to 0.65 ± 0.1 of starting levels. For Tb927.3.3730 the transfer of either the 5′ or the 3′ UTR to the reporter resulted in a 1.6 ± 0.1 fold or 1.4 ± 0.2-fold increase in mRNA levels, respectively. These increases, if combined, may be sufficient to explain the increase in Tb927.3.3730 levels of 3.95 ± 0.3. For Tb927.11.12100, only the 3′ UTR transferred an increase in mRNA level to the reporter mRNA. The increase in mRNA level, 2.4 ± 0.3-fold, was slightly lower than the 3.4 ± 0.4-fold increase in Tb927.11.12100 mRNA. For the two genes that are downregulated in response to dhh1 DEAD:DQAD expression, Tb927.10.15410 and Tb927.6.2790, the presence of either 5′ UTR in the reporter did not result in any additional downregulation. The presence of either 3′ UTR in the reporter resulted in a decrease in mRNA levels, 0.27 ± 0.1 fold for the Tb927.10.15410 3′UTR and 0.49 ± 0.1 fold for Tb927.6.2790 3′UTR, but the reduction was less than seen for the wild type mRNAs and only significant for Tb927.10.15410 (Fig. 3 B,C).
In conclusion, the UTRs of the dhh1 DEAD:DQAD target genes can transfer the response to a reporter gene. In three cases, the 3′UTR acts alone, but for the one gene with the long 5′ UTR (Tb927.3.3730, ), both UTRs contribute. For some genes, the effect of the 3′ UTR on the reporter genes is significantly lower than on the dhh1 target gene; the most likely explanation is that the elements in the reporter mRNA have an opposing effect on mRNA stability.
Mutation of an AU-rich instability element in the mRNA 3′ UTR prevents the dhh1 response and results in mRNA stabilization
mRNAs that are downregulated in response to dhh1 DEAD:DQAD expression may simply become unstable as a secondary effect of repression of translation. mRNAs with an increase in stability, in contrast, must result from some specific effect of the helicase. For this reason, one of the upregulated mRNAs, Tb927.11.12100, was chosen for a more detailed analysis. Systematic 3′ UTR truncation experiments based on the reporter system described above located a necessary and sufficient element for a dhh1 DEAD:DQAD response in the first 318 nucleotides of the 3′UTR downstream of the stop codon (data not shown). This 318 nucleotide element contained several U-rich and AU-rich stretches and such motifs have been previously shown to mediate instability to trypanosome mRNAs.Citation42,Citation43
To test whether these AU-rich elements were responsible for the dhh1 response, the endogenous locus of Tb927.11.12100 was modified in a cell line with an inducible dhh1 DEAD:DQAD transgene and one allele of Tb927.11.12100 deleted. The remaining Tb927.11.12100 allele was mutated in the context of the endogenous 5′ UTR, ORF and polyadenylation site by targeted replacement of the sequence between the Tb927.11.12100 and downstream ORFs. The resultant transgenes were wild type with the exception of sequences between two unique SwaI and BstEII restriction enzyme sites in the 3′ UTR that contained the majority of the necessary and sufficient element (246 nucleotides). Four different cell lines were generated, one with the wild type sequences, one with the AU-rich sequences mutated to decrease the percentage AU and remove homopolymer U runs, one with the AU-rich sequences deleted and one that lacked the entire 236 nucleotides between the two restriction enzyme sites (). The levels of Tb927.11.12100 mRNAs were analyzed by quantitative northern blot, using a Tb927.11.12100 ORF probe, for two independent clones of each transgenic cell line before and after dhh1 DEAD:DQAD induction. RNA from BSF and PCF wild type cells served as control and showed the expected developmental regulation.
Figure 4. Mutagenesis of the AU-rich element of Tb927.11.12100 abolishes the response to dhh1 DEAD:DQAD induction. (A) One allele of Tb927.11.12100 was deleted and the second modified as shown. Cell lines were made in which the various altered sequences shown were introduced between the SwaI and BstEII sites. U-rich sequences are shown in blue and mutations in red. (B) northern blots of total RNA before and after induction of dhh1 DEAD:DQAD (0 or 24 h TET) from cell lines with the indicated modifications to one allele of Tb927.11.12100 and with the second allele deleted. A probe derived from the Tb927.11.12100 ORF was used for probing. Blots and quantifications are shown for two independent clones of each cell line. RNA from wild type BSF and PCF cells are included as a comparison and rRNA served as loading control.
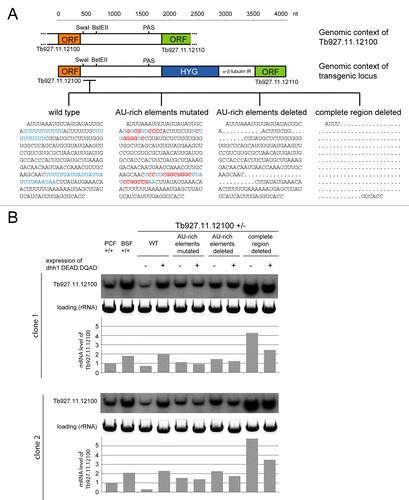
Any interference with AU-rich sequences increased the level of mRNA before dhh1 DEAD:DQAD induction, indicating a destabilizing function. The more the AU-rich element was altered, the greater the stabilization of the mRNA: there was a ~2-fold increase in mRNA level with the AU-rich sequences mutated, a 2- to 5-fold increase when the sequences were deleted and a more than a 5-fold increase when the entire region was deleted. In addition, any interference with the AU-rich element completely prevented the increase in mRNA level that occurs with the wild type after dhh1 DEAD:DQAD induction, indicating that the AU-rich element is required for the response. Instead, mRNA levels were slightly reduced by dhh1 DEAD:DQAD expression; this decrease occurs for the majority of mRNAs and is likely a consequence of the general repression of translation caused by the dhh1 DEAD:DQAD expression.Citation40 Taken together, the data show that the two small AU-rich elements act as instability elements and are necessary for the mRNA stabilization by the dhh1 mutant. It is likely that the 246 nucleotide sequence contains additional elements that mediate mRNA instability, because the deletion of the entire region caused more mRNA stabilization than the deletion of just the AU-rich elements. However, these were not involved in the dhh1 response and therefore not further examined.
The AU-rich element does not mediate developmental regulation
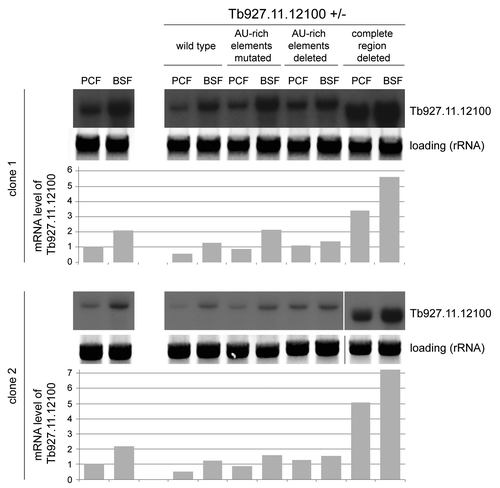
Many of the mRNAs stabilized by the expression of the dhh1 mutant are developmentally regulated; this includes Tb927.11.12100.Citation40 The effect of the mutations in the AU-rich element on the developmental regulation of Tb927.11.12100 was tested by introducing the same transgenes into bloodstream form trypanosomes with a deletion of one allele of Tb927.11.12100. The steady-state levels of Tb927.11.12100 mRNA from the cells with wild type 3′UTR as well as from the three mutant UTRs was analyzed by northern blot and compared with mRNAs from the analogous procyclic mutants (). The developmental regulation of the Tb927.11.12100 mRNA remained unaltered, when the AU-rich motif was mutated or when the entire region was deleted, but was slightly reduced when the AU-rich motif was deleted. The data show that the AU-rich element is not responsible for developmental regulation and that at least for this dhh1 target gene, the cis-elements responsible for stabilization following dhh1 DEAD:DQAD expression and for developmental regulation are distinct.
Figure 5. Mutagenesis of the AU-rich element of Tb927.11.12100 does not affect developmental regulation. Northern blots of total RNA from BSF and PCF cell lines with the indicated modifications to one allele of Tb927.11.12100 (compare with ) and with the second allele deleted were probed for the Tb927.11.12100 ORF. Blots and quantifications are shown for two independent clones of each cell line. RNA from wild type BSF and PCF cells are included as a comparison and rRNA served as loading control. The differences in signal strength between the clones result from different exposure times.
Discussion
DHH1 is best known as a repressor of translation and/or decappingCitation16-Citation21 and binds mRNAs independently of sequence.Citation5,Citation7 However, DHH1 can also selectively affect individual mRNAs. In trypanosomes, DHH1 overexpression or expression of a dhh1 DEAD:DQAD mutant, selectively stabilizes a small set of transcripts while another set of transcripts decreases in level.Citation40 The observed changes in mRNA stability are likely caused by an increase in DHH1 interaction with mRNA, caused by either DHH1 overexpression or more strongly by the expression of dhh1 DEAD:DQAD, an ATPase mutant that is probably impaired in mRNA release,Citation3,Citation4 but not in mRNA unwinding.Citation5 This study aimed to gain insight into the mechanism of selective mRNA stabilization / destabilization by DHH1. Four mRNAs were analyzed, the response to dhh1 DEAD:DQAD expression was mediated by the 3′ UTR for three and by both 5′ and 3′ UTRs for one. One of the upregulated mRNAs was analyzed in detail, the responsible sequences were present in an AU-rich instability cis-element located in the first 318 of the 1200 nucleotide 3′UTR. This is the first report of an AU-rich instability element mediating an increase, rather than a decrease, in mRNA stability on increased dhh1 activity.
Essentially, four different mechanisms could explain the selective effects of DHH1 on individual mRNAs. The most obvious mechanism is the recruitment of DHH1 to mRNAs via RNA binding proteins that specifically recognize mRNA targets via cis-acting elements. At least four different RNA binding proteins have been described in other organisms to directly bind to DHH1 and this way causing mRNA targets to be removed from translation and/or degraded.Citation11,Citation22-Citation25,Citation44 These include two proteins that bind to AU-rich elements, TTPCitation24 and CTH2.Citation25 Our data do not support such a recruitment model for the action of DHH1 for the Tb927.11.12100 mRNA investigated here. The AU-rich instability element in the 3′ UTR of Tb927.11.12100 mediates an increase in mRNA stability on increased DHH1 occupancy not a decrease or translational repression, as observed in all the reported cases of DHH1 recruitment via RNA binding proteins. A difference in DHH1 function is unlikely to account for these differences, as both the protein sequence and role in translational repression are conserved.Citation40 Trypanosome DHH1 has neither N- nor C-terminal extensions that could account for organism specific interactions with RNA binding proteins, such as was for instance shown for the yeast DHH1 binding to RBP1.Citation11 In a related species, T. cruzi, DHH1 co-purified PUF6 protein, however, it is unclear whether this binding was direct and, PUF6 appears to destabilize its target mRNAs.Citation45
A second possible mechanism was recently suggested by Sweet et al.Citation21: DHH1 promotes decapping by slowing ribosome movement, essentially indirect recruitment of DHH1 by nucleotide sequence. This model was supported by a DHH1-dependent decrease in stability of the PGK1 mRNA on the introduction of rare codons. Here, the response of mRNAs that contained the same PAC ORF but differed in the UTRs to dhh1 DEAD:DQAD expression was determined. In all cases, the response to dhh1, including both up- and downregulation, was entirely mediated by the UTRs. The independence of the response from the identity of the ORF indicates that the ribosome slowing mechanism is not operating here.
A third mechanism could be that DHH1 selectively binds to individual mRNAs. However, there is currently no evidence for any sequence specificity of DHH1Citation5,Citation7; the one study that showed protection of maternal mRNAs by the C. elegans DHH1 ortholog, CGH-1, did not show direct RNA binding. No common cis-acting element in the trypanosome DHH1 target mRNAs could be readily identified and DHH1 is not associated with polysomes in T. cruziCitation46 or T. brucei.Citation40 A recent study analyzed all transcripts stabilized by the DHH1 overexpression and expression of dhh1 DEAD:DQAD for enrichment of both sequence-based and structure-based 3′UTR elements that were also conserved among different trypanosome species.Citation47 There was an enrichment of the AU-rich element motif AUUUAUU at DHH1 overexpression but not at expression of mutant dhh1.Citation47 However, this motif is not present within the 3′ UTR of the DHH1 target gene analyzed in this work.
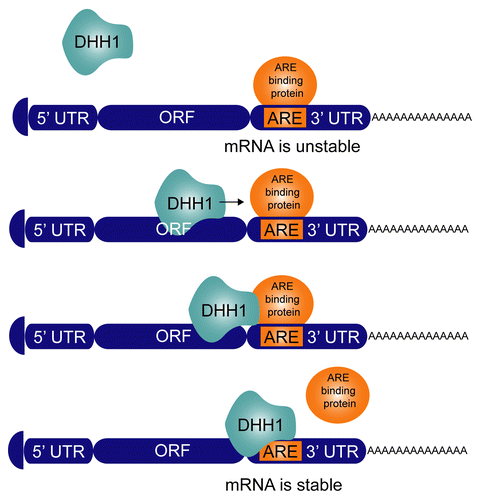
A fourth, novel model for the action of trypanosome DHH1 on individual mRNAs is presented in . In this model, the movement of DHH1 along an mRNA displaces complexes assembled on RNA-binding proteins and once DHH1 has passed, a complex can reassemble. A result of this process is that DHH1 ensures that any mRNA samples the cytoplasmic environment of regulatory proteins at regular intervals. In this context it is worth noting that trypanosomes contain several DHH1 molecules for each mRNA. An increase in DHH1 activity, either through overexpression of wild type, or through increased occupancy as may be the case for dhh1 DEAD:DQAD, will alter the dynamics of mRNP complexes, probably decreasing their stability. This way it could either increase or decrease mRNA stability, depending on whether the regulatory protein had a stabilizing or destabilizing function.
Figure 6. Model for DHH1 action. The fate of an mRNA depends on the binding of trans factors to cis-elements. In this case, ARE-binding proteins bind to AREs in the 3′UTR and result in a short half-life for the mRNA. RNA helicases, in particular DHH1, routinely disrupt these mRNP complexes to facilitate a dynamic interaction between the mRNA and regulatory factors in the cytoplasm. Increase in net DHH1 activity, either by overexpression or by expression of the dhh1 DEAD:DQAD mutant reduces or eliminates complex formation and in this case stabilizes the mRNA.
If a change in DHH1 helicase activity contributes to the regulation of gene expression, how is such a change achieved in the cell? Possibilities are a change in expression level, a change in activity by post-translational modifications or the binding of a regulatory protein or a reduction in available protein levels by recruitment of DHH1 away from translation, for instance to P-bodies. It currently remains unclear, whether any such changes occur, in particular whether they occur during the trypanosome life-cycle.
The action of dhh1 DEAD:DQAD on Tb927.11.12100 mRNA was mediated by an ARE and mutation or deletion of the ARE resulted in equivalent stabilization of the mRNA in both life cycle stages tested. In trypanosomatid species, AREs can be involved in life cycle stage specific mRNA stabilityCitation42,Citation47-Citation49; at least in one case mediated by stage specific ARE-binding proteins.Citation50 However, here the ARE acts as a regulator of steady-state mRNA levels but is not necessary for the developmentally regulated differences. Presumably, a second mechanism, either stabilizing or destabilizing acts in addition.
This is the first report of an AU-rich cis-acting element that is responsible for DHH1 induced mRNA stabilization. Our proposed mode of action, a decrease in the stability of mRNP complexes containing regulatory mRNA binding proteins, provides an explanation, of how a switch in gene expression could be achieved by a change in the available helicase activity. Such a mechanism of helicase action may not be restricted to DHH1 or to trypanosomes. The need to switch from one set of gene to another is a frequent need in biology, for instance during development or in stress situations.
Material and Methods
Plasmids
All constructs were made using standard amplification and cloning procedures. The sequence of all amplified DNAs was verified.
Open reading frame replacement
Plasmids containing a puromycin acetyl transferase (PAC) ORF flanked by 5′ and 3′ sequences immediately adjacent to the target ORFs were constructed. The lengths of the flanking sequences were: 530, 1343, and 542 nucleotides for the 5′ sequence and 519, 1606, 1824 nucleotides for the 3′ sequence for Tb927.11.12100, Tb927.10.15410 and Tb927.6.2790 respectively. Before transfection, the plasmid was cut with two restriction enzymes cutting on either side of the replacement cassette.
Fusion of the 5′ UTRs to the eIF4E3 reporter gene (Supplemental Fig. S2B)
The plasmid contained (5′- > 3′): the 456 nt immediately upstream of the eIF4E3 (Tb927.11.11770) ORF, a PAC ORF, the sequence between the stop codon of the gene upstream of the DHH1 target gene and the start codon of the DHH1 target gene and the N-terminal 459 nucleotides of the eIF4E3 ORF. In addition, for three of the dhh1 target genes (Tb927.10.15410, Tb927.6.2790, Tb927.11.12100) an eYFP gene was fused N-terminally to the eIF4E3 ORF to increase the length of the mRNA to enable simultaneous detection of both the wild type and the genetically engineered eIF4E3 allele.
Fusion of the 3′ UTRs to the eIF4E3 reporter gene (Supplemental Fig. S2C)
The plasmids contained (5′- > 3′): the C-terminal 537 nt of the eIF4E3 ORF, the entire non-coding sequence between the dhh1 target gene stop codon and the start codon of the downstream gene, a PAC ORF and the C-terminal 465 nt of the eIF4E3 3′ UTR.
Mutations/Deletions in the AU-rich element of Tb927.11.12100
The construct used for integration into the genome contained (5′- > 3′): the Tb927.11.12100 open reading frame, the entire sequence between the stop codon of Tb927.11.12100 and the start codon of the downstream ORF, a hygromycin resistance ORF, the entire non-coding sequence between the stop codon of the T. brucei α tubulin gene and the start codon of the β tubulin gene and the first 553 nucleotides of the open reading frame of the gene downstream of Tb927.11.12100. Two unique restriction enzymes sites (SwaI/BstEII) were used to either delete part of the Tb927.11.12100 UTR or to insert a mutated version.
Trypanosomes and transgenic cell lines
T. brucei Lister 427 procyclic cells and MITat1.6 bloodstream form trypanosomes were used. The replacement of the ORF and the fusion of the UTRs to the eIF4E3 reporter gene were done in the previously described PTT cell line (Philippe Bastin, Institute Pasteur Paris, France) engineered for inducible expression of dhh1 DEAD:DQAD from pLEW100.Citation40,Citation51 The mutations of the AU-rich element of Tb927.11.12100 were done in the SPR2.1 cell lineCitation52 engineered for inducible expression of dhh1 DEAD:DQAD from p3383.Citation52 All PCF and BSF cell lines with mutation / deletions in the AU-rich region were verified by PCR and sequencing for correct integration of the plasmid.
Quantitative northern blots
Northern blots were done as described previously.Citation53 Phosphorimager analysis was used for quantification and rRNA served as loading control.
Additional material
Download Zip (94 KB)Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors like to thank Nancy Standart for critical reading of the manuscript, Jenny Reed (Cambridge) as well as all members of Zoology I (Würzburg) for great support and Markus Engstler (Würzburg) for providing excellent infrastructure. This work was funded by the Wellcome Trust (085956/Z/08/Z) and by the DFG (grant Kr4017/1–1).
References
- Russell R, Jarmoskaite I, Lambowitz AM. Toward a molecular understanding of RNA remodeling by DEAD-box proteins. RNA Biol 2013; 10:44 - 55; http://dx.doi.org/10.4161/rna.22210; PMID: 22995827
- Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem 2007; 76:23 - 50; http://dx.doi.org/10.1146/annurev.biochem.76.052305.115300; PMID: 17506634
- Liu F, Putnam A, Jankowsky E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci U S A 2008; 105:20209 - 14; http://dx.doi.org/10.1073/pnas.0811115106; PMID: 19088201
- Aregger R, Klostermeier D. The DEAD box helicase YxiN maintains a closed conformation during ATP hydrolysis. Biochemistry 2009; 48:10679 - 81; http://dx.doi.org/10.1021/bi901278p; PMID: 19839642
- Ernoult-Lange M, Baconnais S, Harper M, Minshall N, Souquere S, Boudier T, Bénard M, Andrey P, Pierron G, Kress M, et al. Multiple binding of repressed mRNAs by the P-body protein Rck/p54. RNA 2012; 18:1702 - 15; http://dx.doi.org/10.1261/rna.034314.112; PMID: 22836354
- Presnyak V, Coller J. The DHH1/RCKp54 family of helicases: an ancient family of proteins that promote translational silencing. Biochim Biophys Acta 2013; 1829:817-23.
- Mitchell SF, Jain S, She M, Parker R. Global analysis of yeast mRNPs. Nat Struct Mol Biol 2013; 20:127 - 33; http://dx.doi.org/10.1038/nsmb.2468; PMID: 23222640
- Ladomery M, Wade E, Sommerville J. Xp54, the Xenopus homologue of human RNA helicase p54, is an integral component of stored mRNP particles in oocytes. Nucleic Acids Res 1997; 25:965 - 73; http://dx.doi.org/10.1093/nar/25.5.965; PMID: 9023105
- Dutta A, Zheng S, Jain D, Cameron CE, Reese JC. Intermolecular interactions within the abundant DEAD-box protein Dhh1 regulate its activity in vivo. J Biol Chem 2011; 286:27454 - 70; http://dx.doi.org/10.1074/jbc.M111.220251; PMID: 21642421
- Weston A, Sommerville J. Xp54 and related (DDX6-like) RNA helicases: roles in messenger RNP assembly, translation regulation and RNA degradation. Nucleic Acids Res 2006; 34:3082 - 94; http://dx.doi.org/10.1093/nar/gkl409; PMID: 16769775
- Chang LC, Lee FJ. The RNA helicase Dhh1p cooperates with Rbp1p to promote porin mRNA decay via its non-conserved C-terminal domain. Nucleic Acids Res 2012; 40:1331 - 44; http://dx.doi.org/10.1093/nar/gkr803; PMID: 21998293
- Drummond SP, Hildyard J, Firczuk H, Reamtong O, Li N, Kannambath S, Claydon AJ, Beynon RJ, Eyers CE, McCarthy JE. Diauxic shift-dependent relocalization of decapping activators Dhh1 and Pat1 to polysomal complexes. Nucleic Acids Res 2011; 39:7764 - 74; http://dx.doi.org/10.1093/nar/gkr474; PMID: 21712243
- Nissan T, Rajyaguru P, She M, Song H, Parker R. Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol Cell 2010; 39:773 - 83; http://dx.doi.org/10.1016/j.molcel.2010.08.025; PMID: 20832728
- Hata H, Mitsui H, Liu H, Bai Y, Denis CL, Shimizu Y, Sakai A. Dhh1p, a putative RNA helicase, associates with the general transcription factors Pop2p and Ccr4p from Saccharomyces cerevisiae. Genetics 1998; 148:571 - 9; PMID: 9504907
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 2000; 403:623 - 7; http://dx.doi.org/10.1038/35001009; PMID: 10688190
- Fischer N, Weis K. The DEAD box protein Dhh1 stimulates the decapping enzyme Dcp1. EMBO J 2002; 21:2788 - 97; http://dx.doi.org/10.1093/emboj/21.11.2788; PMID: 12032091
- Coller JM, Tucker M, Sheth U, Valencia-Sanchez MA, Parker R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA 2001; 7:1717 - 27; http://dx.doi.org/10.1017/S135583820101994X; PMID: 11780629
- Minshall N, Standart N. The active form of Xp54 RNA helicase in translational repression is an RNA-mediated oligomer. Nucleic Acids Res 2004; 32:1325 - 34; http://dx.doi.org/10.1093/nar/gkh303; PMID: 14982957
- Minshall N, Kress M, Weil D, Standart N. Role of p54 RNA helicase activity and its C-terminal domain in translational repression, P-body localization and assembly. Mol Biol Cell 2009; 20:2464 - 72; http://dx.doi.org/10.1091/mbc.E09-01-0035; PMID: 19297524
- Carroll JS, Munchel SE, Weis K. The DExD/H box ATPase Dhh1 functions in translational repression, mRNA decay, and processing body dynamics. J Cell Biol 2011; 194:527 - 37; http://dx.doi.org/10.1083/jcb.201007151; PMID: 21844211
- Sweet T, Kovalak C, Coller J. The DEAD-box protein Dhh1 promotes decapping by slowing ribosome movement. PLoS Biol 2012; 10:e1001342; http://dx.doi.org/10.1371/journal.pbio.1001342; PMID: 22719226
- Minshall N, Reiter MH, Weil D, Standart N. CPEB interacts with an ovary-specific eIF4E and 4E-T in early Xenopus oocytes. J Biol Chem 2007; 282:37389 - 401; http://dx.doi.org/10.1074/jbc.M704629200; PMID: 17942399
- Jang L-T, Buu L-M, Lee F-JS. Determinants of Rbp1p localization in specific cytoplasmic mRNA-processing foci, P-bodies. J Biol Chem 2006; 281:29379 - 90; http://dx.doi.org/10.1074/jbc.M601573200; PMID: 16885161
- Qi MY, Wang ZZ, Zhang Z, Shao Q, Zeng A, Li XQ, Li WQ, Wang C, Tian FJ, Li Q, et al. AU-rich-element-dependent translation repression requires the cooperation of tristetraprolin and RCK/P54. Mol Cell Biol 2012; 32:913 - 28; http://dx.doi.org/10.1128/MCB.05340-11; PMID: 22203041
- Pedro-Segura E, Vergara SV, Rodríguez-Navarro S, Parker R, Thiele DJ, Puig S. The Cth2 ARE-binding protein recruits the Dhh1 helicase to promote the decay of succinate dehydrogenase SDH4 mRNA in response to iron deficiency. J Biol Chem 2008; 283:28527 - 35; http://dx.doi.org/10.1074/jbc.M804910200; PMID: 18715869
- Kabani S, Fenn K, Ross A, Ivens A, Smith TK, Ghazal P, Matthews K. Genome-wide expression profiling of in vivo-derived bloodstream parasite stages and dynamic analysis of mRNA alterations during synchronous differentiation in Trypanosoma brucei. BMC Genomics 2009; 10:427; http://dx.doi.org/10.1186/1471-2164-10-427; PMID: 19747379
- Siegel TN, Hekstra DR, Wang X, Dewell S, Cross GAM. Genome-wide analysis of mRNA abundance in two life-cycle stages of Trypanosoma brucei and identification of splicing and polyadenylation sites. Nucleic Acids Res 2010; 38:4946 - 57; http://dx.doi.org/10.1093/nar/gkq237; PMID: 20385579
- Jensen BC, Sivam D, Kifer CT, Myler PJ, Parsons M. Widespread variation in transcript abundance within and across developmental stages of Trypanosoma brucei. BMC Genomics 2009; 10:482; http://dx.doi.org/10.1186/1471-2164-10-482; PMID: 19840382
- Queiroz R, Benz C, Fellenberg K, Hoheisel JD, Clayton C. Transcriptome analysis of differentiating trypanosomes reveals the existence of multiple post-transcriptional regulons. BMC Genomics 2009; 10:495; http://dx.doi.org/10.1186/1471-2164-10-495; PMID: 19857263
- Campbell DA, Thornton DA, Boothroyd JC. Apparent discontinuous transcription of Trypanosoma brucei variant surface antigen genes. Nature 1984; 311:350 - 5; http://dx.doi.org/10.1038/311350a0; PMID: 6090933
- LeBowitz JH, Smith HQ, Rusche L, Beverley SM. Coupling of poly(A) site selection and trans-splicing in Leishmania. Genes Dev 1993; 7:996 - 1007; http://dx.doi.org/10.1101/gad.7.6.996; PMID: 8504937
- Ullu E, Matthews KR, Tschudi C. Temporal order of RNA-processing reactions in trypanosomes: rapid trans splicing precedes polyadenylation of newly synthesized tubulin transcripts. Mol Cell Biol 1993; 13:720 - 5; PMID: 8417363
- Matthews KR, Tschudi C, Ullu E. A common pyrimidine-rich motif governs trans-splicing and polyadenylation of tubulin polycistronic pre-mRNA in trypanosomes. Genes Dev 1994; 8:491 - 501; http://dx.doi.org/10.1101/gad.8.4.491; PMID: 7907303
- Günzl A, Bruderer T, Laufer G, Schimanski B, Tu L-C, Chung H-M, Lee P-T, Lee MG-S. RNA polymerase I transcribes procyclin genes and variant surface glycoprotein gene expression sites in Trypanosoma brucei. Eukaryot Cell 2003; 2:542 - 51; http://dx.doi.org/10.1128/EC.2.3.542-551.2003; PMID: 12796299
- Kramer S, Carrington M. Trans-acting proteins regulating mRNA maturation, stability and translation in trypanosomatids. Trends Parasitol 2011; 27:23 - 30; http://dx.doi.org/10.1016/j.pt.2010.06.011; PMID: 20609625
- Kramer S. Developmental regulation of gene expression in the absence of transcriptional control: the case of kinetoplastids. Mol Biochem Parasitol 2012; 181:61 - 72; http://dx.doi.org/10.1016/j.molbiopara.2011.10.002; PMID: 22019385
- Araújo PR, Teixeira SM. Regulatory elements involved in the post-transcriptional control of stage-specific gene expression in Trypanosoma cruzi: a review. Mem Inst Oswaldo Cruz 2011; 106:257 - 66; PMID: 21655811
- Milone J, Wilusz J, Bellofatto V. Identification of mRNA decapping activities and an ARE-regulated 3′ to 5′ exonuclease activity in trypanosome extracts. Nucleic Acids Res 2002; 30:4040 - 50; http://dx.doi.org/10.1093/nar/gkf521; PMID: 12235388
- Kramer S, Queiroz R, Ellis L, Hoheisel JD, Clayton C, Carrington M. The RNA helicase DHH1 is central to the correct expression of many developmentally regulated mRNAs in trypanosomes. J Cell Sci 2010; 123:699 - 711; http://dx.doi.org/10.1242/jcs.058511; PMID: 20124414
- Kolev NG, Franklin JB, Carmi S, Shi H, Michaeli S, Tschudi C. The transcriptome of the human pathogen Trypanosoma brucei at single-nucleotide resolution. PLoS Pathog 2010; 6:e1001090; http://dx.doi.org/10.1371/journal.ppat.1001090; PMID: 20838601
- Di Noia JM, D’Orso I, Sánchez DO, Frasch AC. AU-rich elements in the 3′-untranslated region of a new mucin-type gene family of Trypanosoma cruzi confers mRNA instability and modulates translation efficiency. J Biol Chem 2000; 275:10218 - 27; http://dx.doi.org/10.1074/jbc.275.14.10218; PMID: 10744707
- Quijada L, Guerra-Giraldez C, Drozdz M, Hartmann C, Irmer H, Ben-Dov C, Cristodero M, Ding M, Clayton C. Expression of the human RNA-binding protein HuR in Trypanosoma brucei increases the abundance of mRNAs containing AU-rich regulatory elements. Nucleic Acids Res 2002; 30:4414 - 24; http://dx.doi.org/10.1093/nar/gkf577; PMID: 12384588
- Standart N, Minshall N. Translational control in early development: CPEB, P-bodies and germinal granules. Biochem Soc Trans 2008; 36:671 - 6; http://dx.doi.org/10.1042/BST0360671; PMID: 18631138
- Dallagiovanna B, Correa A, Probst CM, Holetz F, Smircich P, de Aguiar AM, Mansur F, da Silva CV, Mortara RA, Garat B, et al. Functional genomic characterization of mRNAs associated with TcPUF6, a pumilio-like protein from Trypanosoma cruzi. J Biol Chem 2008; 283:8266 - 73; http://dx.doi.org/10.1074/jbc.M703097200; PMID: 18056709
- Holetz FB, Correa A, Avila AR, Nakamura CV, Krieger MA, Goldenberg S. Evidence of P-body-like structures in Trypanosoma cruzi. Biochem Biophys Res Commun 2007; 356:1062 - 7; http://dx.doi.org/10.1016/j.bbrc.2007.03.104; PMID: 17399688
- Najafabadi HS, Lu Z, MacPherson C, Mehta V, Adoue V, Pastinen T, Salavati R. Global identification of conserved post-transcriptional regulatory programs in trypanosomatids. Nucleic Acids Res 2013; 41:8591 - 600; http://dx.doi.org/10.1093/nar/gkt647; PMID: 23877242
- Hotz HR, Hartmann C, Huober K, Hug M, Clayton C. Mechanisms of developmental regulation in Trypanosoma brucei: a polypyrimidine tract in the 3′-untranslated region of a surface protein mRNA affects RNA abundance and translation. Nucleic Acids Res 1997; 25:3017 - 26; http://dx.doi.org/10.1093/nar/25.15.3017; PMID: 9224601
- Mayho M, Fenn K, Craddy P, Crosthwaite S, Matthews K. Post-transcriptional control of nuclear-encoded cytochrome oxidase subunits in Trypanosoma brucei: evidence for genome-wide conservation of life-cycle stage-specific regulatory elements. Nucleic Acids Res 2006; 34:5312 - 24; http://dx.doi.org/10.1093/nar/gkl598; PMID: 17012283
- D’Orso I, Frasch AC. TcUBP-1, a developmentally regulated U-rich RNA-binding protein involved in selective mRNA destabilization in trypanosomes. J Biol Chem 2001; 276:34801 - 9; http://dx.doi.org/10.1074/jbc.M102120200; PMID: 11435421
- Wirtz E, Leal S, Ochatt C, Cross GA. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol 1999; 99:89 - 101; http://dx.doi.org/10.1016/S0166-6851(99)00002-X; PMID: 10215027
- Sunter J, Wickstead B, Gull K, Carrington M. A new generation of T7 RNA polymerase-independent inducible expression plasmids for Trypanosoma brucei. PLoS One 2012; 7:e35167; http://dx.doi.org/10.1371/journal.pone.0035167; PMID: 22511983
- Kramer S, Queiroz R, Ellis L, Webb H, Hoheisel JD, Clayton C, Carrington M. Heat shock causes a decrease in polysomes and the appearance of stress granules in trypanosomes independently of eIF2(alpha) phosphorylation at Thr169. J Cell Sci 2008; 121:3002 - 14; http://dx.doi.org/10.1242/jcs.031823; PMID: 18713834
