Abstract
During apoptosis, activated caspases cleave the translation initiation factor eIF4G. This cleavage disrupts cap-dependent mRNA translation initiation within the cell. However, a specific subset of mRNAs can still be recruited for protein synthesis in a cap-independent manner by the residual initiation machinery. Many of these mRNAs, including cell death related mRNAs, contain internal ribosome entry sites (IRESes) that promote their enhanced translation during apoptosis. Still other mRNAs have little dependence on the cap recognition mechanism. The expression of the encoded proteins, both anti- and pro-apoptotic, allows for an initial period of attempted cell survival, then commitment to cell death when damage is extensive. In this study we address the translational regulation of the stress and apoptosis-related mRNAs in C. elegans: BiP (hsp-3) (hsp-4), Hif-1 (hif-1), p53 (cep-1), Bcl-2 (ced-9) and Apaf-1 (ced-4). Altered translational efficiency of these messages was observed upon depletion of cap-dependent translation and induction of apoptosis within the C. elegans gonad. Our findings suggest a physiological link between the cap-independent mechanism and the enhanced translation of hsp-3 and ced-9. This increase in the efficiency of translation may be integral to the stress response during the induction of physiological apoptosis.
Introduction
Stem cell fate decisions in the C. elegans germ line are regulated primarily by mRNA translational control.Citation1 All germ cells entering meiosis experience chromosome condensation that correlates with suppressed transcription. Therefore, changes in protein expression leading to cell fate decisions result from translational regulation of stored mRNAs. One vital fate decision for developing germ cells is the decision between apoptosis and continued maturation. During C. elegans oogenesis, nearly half of the progenitor cell population is naturally fated for death before reaching maturity.Citation2 These cells contribute cytoplasmic contents (mRNAs, ribosomes, mitochondria) and nutrients to the maturing sibling oocytes.Citation2 The dying oocytes are thought to perform a similar role to nurse cells in the ovaries of higher animals. Thus, oogenesis in worms provides a versatile model to study the role of protein synthesis regulation in an unusual cell fate decision to differentiate naturally toward apoptosis.
The translation initiation complex selects mRNAs for protein synthesis. Initiation factor eIF4G provides a scaffold for the assembly of other initiation factors and ribosomes on recruited mRNAs.Citation3,Citation4 In mammals there are three eIF4G isoforms: eIF4GI, eIF4GII and p97.Citation5,Citation6 eIF4GI and eIF4GII contain binding domains for eIF4E, which binds to the mRNA cap, eIF3 and eIF4A, which associate with the 40S subunit of the ribosome, and poly(A) binding proteins (PABP).Citation3,Citation4,Citation7 These domains promote the recruitment of mRNA to the ribosome for translation.Citation8 Caspase-3 cleaves eIF4GI and eIF4GII during apoptosis, removing their eIF4E and PABP binding domains preventing their involvement in translation initiation.Citation7,Citation9-Citation12 eIF4GII becomes degraded and plays no further role in protein synthesis.Citation13 However, translation initiation still occurs on a select subset of mRNAs via cap-independent translation.Citation14 Cleaved eIF4GI, and its cap-independent paralog, p97, are known to bind directly to mRNA, often via internal ribosome entry sites (IRESes).Citation12,Citation14 This association allows cleaved eIF4GI and p97 to recruit a specific population of mRNAs for translation during stress.Citation12,Citation14 We have previously shown in C. elegans that CED-3 (caspase) cleavage of IFG-1 p170 (eIF4GI) during apoptosis removes its cap-associated domain much like the mammalian ortholog.Citation15 After cleavage, cap-independent initiation allows cleaved IFG-1 p170 (not cap-associated) and IFG-1 p130 (constitutively not cap-associated) to recruit mRNAs for translation.Citation16 Unlike in mammalian cells, only these two eIF4G forms are found in C. elegans and are encoded by a single ifg-1 gene. Thus, we are investigating changes in translation initiation that result from the differential usage of IFG-1 p170, IFG-1 p130 and their caspase cleaved products. IFG-1 cleavage may allow for the differential expression of proteins determining whether the oocyte matures or succumbs to apoptosis. Importantly, we have previously demonstrated that the IFG-1 p170 loss is not merely the cell’s attempt to shut down all protein synthesis, leading to inevitable cell death. Instead, our epistasis experiments revealed that the disruption of cap-dependent translation acts as an upstream effector of cellular suicide acting through the apoptotic (ced) signaling pathway.Citation15 Our model proposes that changes in protein expression, resulting from eIF4G cleavage, are an integral part of apoptotic cell fate decisions.
eIF4G cleavage results in a change in the population of mRNAs being translated. Among these target mRNAs are both pro- and anti-apoptotic mRNAs. mRNAs involved in cell cycle progression or cell growth are typically cap-dependent and thus disadvantaged, halting further growth and proliferation.Citation17 Some mRNAs that encode stress-response proteins continue to translate cap-independently via IRESes. Such proteins provide the cell with an opportunity to recover from an apoptotic fate.Citation18 An example of an anti-apoptotic protein whose mRNA translates via an IRES is Binding immunoglobulin Protein (BiP).Citation19 BiP is a chaperone protein located in the endoplasmic reticulum (ER) that facilitates protein folding and promotes recovery from heat shock and ER stress.Citation20 Another example mRNA, Bcl-2, is also translated via an IRES and prevents apoptosome formation.Citation21 The enrichment of anti-apoptotic proteins such as BiP and Bcl-2 during stress conditions presents cells with the means to recover and escape the apoptotic fate. Conversely, a second subset of mRNAs containing IRESes encode apoptotic proteins such as Apaf-1.Citation22 Apaf-1 drives further cell death by promoting apoptosome formation resulting in caspase activation.Citation22 Dynamic regulation of cap-independent translation of pro- and anti-apoptotic mRNAs may cause protein expression patterns that determine the germ cell’s differentiation decision.
Our research addresses the protein synthetic mechanism regulating germ cell fates in the C. elegans model system. The induction of cap-independent translation and its involvement in the natural apoptotic cell fate decision has yet to be studied in a natively differentiating cell lineage. In this study we address the physiological translational control of cap-independent mRNAs. Importantly, our findings show that regulation of some apoptotic mRNAs in this whole organism system differs from observations in cultured mammalian cells. Cap-independent conditions enhance the translational efficiency of some, but not all, C. elegans homologs of known mammalian IRES-containing mRNAs. At least one mRNA that lacks a 5′ IRES (hsp-3) is up-regulated by cap-independent translation. Moreover, selective translation in germ cells differs from the cap-independent translation observed in cultured mammalian cells responding to toxic treatments such as chemotherapy agents and hypoxia.
Results
A splicing defect increases the proportion of non-cap-associated (p130) to cap-associated (p170) IFG-1 isoforms
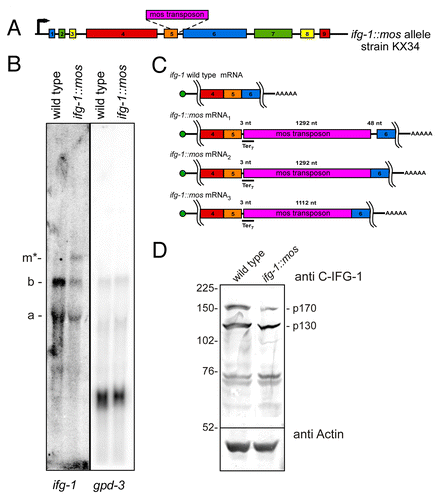
Caspase-mediated cleavage of eIF4GI (IFG-1) during apoptosis increases the ratio of the non-cap-associated IFG-1 p130-like cleavage products to the cap-associated p170.Citation15 This change in ratio may decrease the efficiency of translation of cap-dependent mRNAs and enhance the translation of cap-independent mRNAs. In order to study how this change in ratio of IFG-1 isoforms alters the translation of specific mRNAs in vivo, we developed a mutant strain of C. elegans with a 2.5-fold increase in the p130:p170 IFG-1 ratio (). The change in isoform proportion is caused by a splicing defect in ifg-1 intron 5 that results from a Mos transposon insertion at an intron-exon junction (). Mis-spliced ifg-1::mos mRNAs retain most or all of the Mos transposon sequence (). These mRNAs also have multiple termination codons in all reading frames that will result in nonfunctional truncated N-terminal IFG-1 upon expression (). Truncated proteins are not detected by western blot and are likely degraded ( and data not shown). A small proportion of ifg-1 p170 mRNA (34% of total) properly splices out the Mos sequence (). Properly spliced p130 and p170 mRNAs are translated into functional proteins. However, the overall protein abundance of both IFG-1 isoforms is diminished by 30% (). Western blotting to detect both the p170 and p130 shows that loss of p170 protein expression (to 38% of wild type levels) accounts for nearly all of the decrease. This reduction is likely due to the more rapid turnover of N-terminally extended eIF4G forms and was confirmed using an N-terminal IFG-1 antibody (data not shown).Citation8 As a viable mutant worm strain, ifg-1::mos worms are therefore an appropriate whole animal model in which to address how suppression of cap-associated p170 alters the translational efficiency of specific mRNAs.
Figure 1. A splicing defect increases the proportion of non-cap-associated (p130) to cap-associated (p170) IFG-1 isoforms. (A) Diagram depicting the Mos transposon insert in the ifg-1 gene. The Mos transposon is inserted at 3 bp downstream of the exon-intron 5 junction. The strain was outcrossed 5 times to ensure the absence of additional mutations.(B) Northern blot hybridization depicts ifg-1 message populations. Northern blotting of wild type mRNA displays mRNAs of 2500 nt (a) and 4500 nt (b) indicative of p130 and p170 variants. To control for RNA loading, the blot was stripped and reprobed for gpd-3 RNA. Full-length p170 mRNA (4500 nt) was reduced by 42% and an additional mRNA variant (m*) at approximately 6000 nt was detected in the ifg-1::mos strain that accounted for 17% of all ifg-1 mRNAs. The ratio of p130:p170 mRNA increased 2-fold as p130 mRNA and the total ifg-1 mRNA amounts changed little relative to gpd-3 mRNA. (C) Diagrams representing the four partially spliced variants of ifg-1 mRNA due to the Mos transposon insertion. These splice variants were characterized by RT-PCR and sequencing. All three Mos-containing mRNAs encode nonsense codons in all three reading frames. (D) Western Blot for IFG-1 using the central antibody that detects both the p170 and p130 isoforms. Wild type worms express equal amounts of each IFG-1 isoform (p130:p170 ratio is 1.2), whereas the ifg-1::mos strain p130 abundance is 3.1-fold higher than p170. IFG-1 p170 consistently shows a much greater reduction, likely due to its instability. In the ifg-1::mos strain the total amount of IFG-1 expression is also reduced 30% in comparison to the wild type strain. Western blotting for actin was used as a loading control.
ifg-1::mos and ced-9ts mutant strains exhibit marked increases in germ line apoptosis
Previous experiments demonstrated that RNAi knockdown of cap-associated p170 induced germ cell apoptosis.Citation16 Therefore, we addressed whether the reduced p170 protein abundance in the ifg-1::mos strain lead to a similar induction of germ cell apoptosis. To observe and count apoptotic corpses in the gonad, ifg-1::mos was crossed into a CED-1::GFP-expressing strain. CED-1::GFP is expressed by somatic sheath cells that surround the germ cells in the gonad. Sheath cells invade and engulf the dying oocyte and CED-1::GFP fluorescence allows for live visualization of this process.Citation23 The progression of germ cell apoptosis was measured by counting GFP-decorated oocytes in synchronized late larval to young adult ifg-1::mos and wild type worms over a 48-h period (). Small numbers of germ cell corpses begin to accumulate in both strains as their gonads mature from the larval (L3) stage to the adult stage. This is followed by a steady and significant increase in apoptotic corpses (to an average of 13–15) in ifg-1::mos hermaphrodites in the young adult stages (24–48 h). By contrast, the steady-state number of apoptotic germ cells levels off at about 6 h after wild type worms reach adulthood. The increase in apoptosis in the ifg-1::mos strain indicates that depleting IFG-1 p170 relative to p130 results in increased germ cell apoptosis.
Figure 2.ifg-1::mos transgenic lines exhibits a marked increase in apoptotic corpses in the germ line and a decrease in fertility. (A) A graph depicting CED-1::GFP decorated corpses in the wild type, ifg-1::mos, and ced-9ts strains over time. There is an initial increase in apoptosis in all of the strains as they mature from the L3 larval stage, in which sperm fills the gonad and there is little to no apoptosis. During the L4 larval stage oocytes begin to progress through the gonad and the level of constitutive apoptosis increases. The level of apoptosis within the adult wild type gonad remains constant over time. However, the levels of apoptosis in the ifg-1::mos and ced-9ts lines continue to increase during the adult stage. *ifg-1::mos significantly different from wild type strain P < 0.05, ** P < 0.001, # ced-9ts significantly differs from the wild type strain P < 0.01, ## P < 0.001, § ifg-1::mos significantly differs from ced-9ts strain P < 0.001. (B and C) Fluorescence and DIC image of wild type, ifg-1::mos (D and E) and ced-9ts (F and G) strains expressing the CED-1::GFP apoptotic marker after 24 h at 25 °C. DIC images confirm normal growth and differentiation of oocytes within the gonads of mutant strains. Fertility and extent of larval/adult development were assayed in the wild type (H) and ifg-1::mos (I) strains at 25 °C. Brood size was quantified and the extent of embryonic, larval and adult development was assessed by visual morphology at 24-h intervals for 96 h following egg laying.
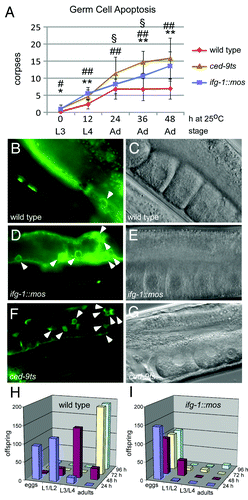
From our previous work and the experiments of others we know that depletion of IFG-1 and CED-9 induces germ cell apoptosis.Citation15,Citation24 Thus, we decided to utilize both of these strains to observe germ line translational control. We compared germ line apoptosis observed in the ifg-1::mos strain to a strain with a temperature sensitive defect in the anti-apoptotic protein CED-9 (Bcl-2). Loss of CED-9 enables apoptosome formation leading to caspase activation and eventual cell death in both somatic and germ cells.Citation25,Citation26 The ced-9ts strain showed a significant increase in apoptotic corpses in the young adult stages (24–48 h) that mirrored the changes observed in the ifg-1::mos strain. Importantly, the activation of the caspase, CED-3, in the ced-9ts strain causes IFG-1 p170 cleavage to a p130-like product, perhaps promoting a similar outcome to the ifg-1::mos allele.Citation15 Thus, the ced-9ts strain similarly allows us to observe how physiological changes in the relative p130:p170 representation may affect translation initiation. The gonads of adult worms of both ifg-1::mos and ced-9ts strains showed visually obvious increases in the prevalence of germ cell corpses (, and ). Importantly, all strains showed normally developed gonads with a typical linear progression of developing germ cells that mature into oocytes (, and ). The strains displayed no other marked morphological defects.
The ifg-1::mos strain displays decreased fertility and embryo viability
The prevalence of apoptosis among germ cells is expected to result in a decrease in the number of viable oocytes reaching maturity. However, it is unclear if surviving oocytes are themselves fully competent to develop as embryos upon fertilization. To correlate the increase in germ cell apoptosis with a decrease in ifg-1::mos fertility, we followed egg laying in wild type and ifg-1::mos hermaphrodites. Simultaneously, we monitored the course of development to address any subsequent developmental defects in the ifg-1::mos hermaphrodites. We found only a modest decrease in the number of total eggs laid by the ifg-1::mos worms at 25 °C ( and ). The wild type strain laid about 228 eggs per hermaphrodite mother, whereas the ifg-1::mos strain laid about 148 eggs. The more significant effect of depleting cap-associated IFG-1 p170 was substantial embryonic lethality and arrest in early larval development. The majority of wild type eggs mature into L3 or L4 larvae by 48 h (). By comparison, less than a third of ifg-1::mos eggs are able to enter larval development and these arrest at larval L1/L2 stages. By 96 h only 11 of those larvae (just under 7% of all eggs laid) reached maturity as adults. In comparison, over 191 wild type eggs (84%) developed into fertile adult offspring (). Diminished fecundity is not an anomaly. Similar results were seen previously with the ifg-1(ok1211) null worms and IFG-1 p170 RNAi. We previously observed both embryonic lethality and L2 larval arrest when IFG-1 function was disrupted.Citation16 L2 larval arrest is the terminal phenotype of ifg-1(ok1211) null worms. Embryonic lethality and the induction of apoptosis were observed upon IFG-1 p170 knockdown due to RNAi. These data suggest that depletion of cap-associated IFG-1 p170 promotes cap-independent translation in the gonad and subsequent apoptosis leading to a corresponding reduction in fertility. Our findings also suggest two other critical developmental steps rely on sufficient IFG-1 activity; progression through embryogenesis and the growth that accompanies early larval development.
hsp-3 mRNA is translated more efficiently when cap-associated IFG-1 p170 is depleted
The ifg-1::mos and ced-9ts strains provided the opportunity to study how changes in the representation of the IFG-1 can affect the recruitment of target mRNAs for translation. Recently, Li, et al. identified an IRES in the 5′ UTR of extended transcripts of the C. elegans hsp-3 mRNA. This IRES allowed hsp-3 mRNA to be translated cap-independently, much like its mammalian homolog BiP.Citation19,Citation27 In human cell lines, the BiP mRNA IRES allows for enhanced BiP expression during heat shock, facilitating cellular recovery by chaperoning protein folding.Citation28 We investigated the translational control of hsp-3 mRNA after depletion of cap-associated IFG-1 p170 and following induction of apoptosis via the ced-9ts/Bcl-2 mutation. Polysome profiling was used to observe changes in translational efficiency of hsp-3 mRNA and the ubiquitously expressed gpd-3 (GAPDH) mRNA as a control. Worm lysates from the wild type, ifg-1::mos, and ced-9ts () strains were fractionated on sucrose gradients to resolve polyribosomal complexes. mRNAs were quantified across the gradient by quantitative real-time PCR (qPCR) to monitor changes in their mRNA distributions that reflect the number of bound ribosomes. The percentage of each specific mRNA represented in each fraction was calculated to determine the translational activity of the target mRNA. Induction of apoptosis was monitored in each strain by visualizing corpses () in order to correlate increased apoptosis with changes in the efficiency of translation of target messages (). As expected, gpd-3 mRNA was translated efficiently in the wild type strain. A modest decrease in the translational efficiency of gpd-3 was detected in the ifg-1::mos strain indicated by the leftward shift in the mRNA distribution. gpd-3, however, still translated pretty efficiently on heavy polysomes (). This modest decrease in translation was not unexpected in the ifg-1::mos strain where reduced levels of both IFG-1 isoforms are found. When the function of Bcl-2 was abrogated in the ced-9ts strain, the distribution of gpd-3 mRNA among polysomes broadened, but the median of the message distribution mirrored that of the wild type strain (). These data demonstrate that the gpd-3 mRNA remains relatively efficiently translated under both conditions that trigger IFG-1 p170 depletion/cleavage and apoptosis.
Figure 3.hsp-3 mRNA is translated more efficiently after the reduction of cap-associated p170 protein abundance. Polysome profiles depicting the continuous monitoring of absorbance at 254nm during sucrose gradient fractionation of worm lysates from wild type (A), ifg-1::mos (B) and ced-9ts (C) strains. These gradients represent 1 of 4 biological replicates of wild type and ced-9ts strains and 1 of 2 biological replicates of the ifg-1::mos strain. The profiles of biological replicates closely resemble one another and confirm the observed changes in translational efficiency. RNA distributions quantified by qPCR in each gradient fraction indicating changes in translational efficiency of gpd-3 in the ifg-1::mos (D) and ced-9ts (E) strains in comparison to the wild type strains. The percent mRNA was quantified by calculating the amount of a specific target mRNA in each fraction then dividing by the total amount of that mRNA across the entire gradient and multiplying by 100. qPCR quantified RNA distributions indicating changes in the translational efficiency of hsp-3 in the ifg-1::mos (F) and ced-9ts (G) strains in comparison to the wild type strain. Changes in translational efficiency of hsp-4 in the ifg-1::mos (H) and ced-9ts (I) strains are indicated by qPCR quantification of gradient fractions. All qPCR results from the ced-9ts strain were confirmed in four biological replicates and ifg-1::mos strain in biological duplicates.
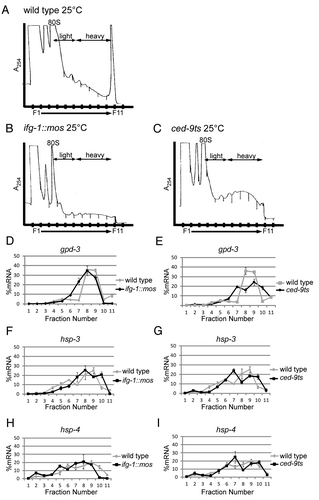
The translational efficiency of hsp-3 was similarly measured by polysome loading to determine if reduced p170 levels, due to the ifg-1 splicing defect (ifg-1::mos) or cleavage during apoptosis (ced-9ts), altered its translation. Both the ifg-1::mos and ced-9ts strains showed consistent, modest increases in hsp-3 mRNA translational efficiency (). This increase is significant due to the overall shift of the mRNA density into the more efficiently translating fractions near the bottom of the gradient. It should be noted that polysome resolution is nonlinear on sucrose gradients. Shifts in position near the top of the gradient (fractions 4–7) represent increases of one or two ribosomes bound. However, equivalent shifts at the bottom of the gradient (fractions 8–11), as are observed for hsp-3 mRNA (especially noting shifts in fraction 10), represent a much larger addition of bound ribosomes and therefore a much greater increase in translational efficiency. hsp-3 mRNA also becomes depleted from light polysomes as additional ribosomes bind and increase the message’s representation in the heavy polysome regions of the gradient. The change in translational efficiency was confirmed in biological replicates (n = 2 for ifg-1::mos; n = 4 for ced-9ts) with similar curves that depicted the same increase in translational efficiency. The authenticity of mRNA sedimentation due to ribosome association was confirmed by pretreatment with EDTA prior to centrifugation to release hsp-3 (and other) mRNAs from ribosomes (data not shown). Most importantly, hsp-3 translates more efficiently under stress and pro-apoptotic conditions that inhibit the translation of most other cellular mRNAs. Increased ribosome association, indicative of enhanced initiation rate, occurred despite decreases in the cap-associated eIF4 complex. This provides a correlation between enhanced hsp-3 translation and cap-independent translation conditions.
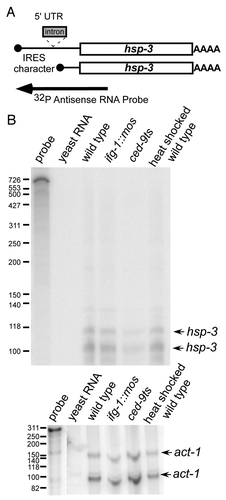
Expressed sequence tags (ESTs) reveal a rare (1 in 300), longer hsp-3 mRNA variant that has IRES characteristics in the extended 5′ UTR. However, the increase in translational efficiency of bulk hsp-3 mRNA most likely is not the result of an IRES in the 5′ UTR. Mapping the 5′ end of the hsp-3 mRNA by RNase protection assay (RPA) using an antisense probe covering all of the mature mRNA forms was performed on RNA from wild type, ifg-1::mos, ced-9ts and wild type temperature stressed worms (). A full-length, 5′-extended mRNA was not detected (). However, two short mRNAs indicative of about a 25 nucleotide 5′ UTR were found. RNA quality was confirmed by RPA using a 3′ UTR act-1 probe. When detecting the translational efficiency of hsp-3 mRNA using qPCR, we used internal primers designed to detect all hsp-3 populations, so it is unlikely the overall translational efficiency would be affected by a minor extended mRNA population, if present. Thus, although hsp-3 is translated more efficiently after the depletion of IFG-1 p170, this increase in translational efficiency is not due in large part to the upstream IRES sequence.
Figure 4. 5′ UTR extended hsp-3 mRNA is not detected. (A) Diagram indicating the coverage of the hsp-3 mRNA probe. The arrow indicates the RPA probe extending upstream from exon 1 and covering all potential mRNA variants. (B) RNase Protection Assays were conducted by hybridizing the 5′ probe to total mRNA in wild type, ced-9ts, ifg-1::mos and wild type heat shocked strains. The arrows indicate the two detected hsp-3 mRNA 5′ variants. Control RNase Protection Assay detecting act-1 mRNA is shown to demonstrate RNA quality.
Another BiP homolog mRNA, hsp-4, has little change in translational efficiency during cap-independent conditions
BiP has two C. elegans homologs HSP-3 and HSP-4.Citation29 Both homologs are localized to the ER as chaperones involved in stress response to heat shock.Citation30 Unlike hsp-3, however, which is constitutively transcribed, hsp-4 mRNA levels are almost undetectable during non-stressed conditions.Citation29 The hsp-4 gene becomes transcriptionally activated with the onset of stress.Citation29 Although hsp-3 and hsp-4 mRNA coding regions share 70% nucleotide identity, hsp-3 has conserved sequences in its 5′ UTR not present in hsp-4.Citation29,Citation30 These sequences are conserved among several distant nematode species and even in rats.Citation30 hsp-4 mRNA also lacks most of the defined IRES region of hsp-3 mRNA. Given the strong homology between these mRNAs but differences in 5′ leader sequences, we sought to determine if hsp-4 mRNA behaves similarly or differently from hsp-3 mRNA upon depletion of IFG-1 p170. We observed that hsp-4 mRNA translational efficiency changed in a way that mirrored changes in gpd-3 mRNA distribution. There was a slight decrease in the efficiency of hsp-4 translation in the ifg-1::mos strain () and no change in ced-9ts worms () relative to wild type. The slight decrease suggests that hsp-3 mRNA has a greater capacity to undergo cap-independent translation, although both messages maintain relatively efficient translation after the knockdown of IFG-1 p170. Thus, while hsp-3 is stress-regulated at the level of translation, hsp-4 may only be regulated by transcriptional control.
Bcl-2 homolog mRNA, ced-9, is translated more efficiently upon cleavage of IFG-1 p170, but not upon genetic depletion
mRNA encoding B-cell lymphoma 2 (Bcl-2), an anti-apoptotic regulator in the apoptotic pathway, is translated cap-independently via an IRES during mammalian apoptosis.Citation31 We assayed the translational control of the C. elegans Bcl-2 homolog, ced-9, by polysome resolution. In wild type worms, ced-9 mRNA is distributed bimodally ( and ). The majority of the mRNA resides in the medium-sized polysomes, while a lesser portion is broadly associated among very light polysomes. Following the induction of apoptosis in ced-9ts mutant worms, ced-9 mRNA from both populations becomes translated more efficiently (). Like hsp-3 mRNA, the heavier peak of ced-9 mRNA moves near the bottom of the gradient from fraction 9 to fraction 10 indicating a substantial increase in the number of bound ribosomes and an increase in the efficiency of translation. Some ced-9 mRNA also mobilizes from the non-translating or very light polysome region into the moderately translating region (from fractions 4–5 into fraction 7). The ced-9ts strain undergoes loss of CED-9 function due to a point mutation in ced-9 mRNA. This mutation did not result in changes in ced-9 mRNA accumulation nor directly enhanced ced-9 mRNA translation. Rather the changes in the efficiency of translation resulted from the induction of apoptosis. Thus, ced-9 in the whole worm was translated similarly to its mammalian homolog during apoptotic stress. Conversely, ced-9 mRNA translation was not enhanced by IFG-1 depletion in the ifg-1::mos strain (). It is possible that the decrease in both IFG-1 p170 and p130 in this strain is suboptimal for ced-9 mRNA initiation. CED-9 has both pro- and anti-apoptotic functions in C. elegans making it an interesting candidate for translational control during apoptosis.Citation32 Overall, these results suggest that cleavage of IFG-1 p170, resulting in increased p130-like products, may differ translationally from genetic IFG-1 depletion, resulting in decreased expression of both p170 and p130.
Figure 5. Other stress and apoptotic mRNAs, ced-9 (Bcl-2), hif-1 (HIF-1), cep-1 (p53) and ced-4 (Apaf-1) have differing abilities to be translated after depletion of cap-associated IFG-1 p170. RNA distributions were quantified by qPCR in wild type, ifg-1::mos, and ced-9ts strains from polysome profiles depicted in . These gradients represent 1 of 4 biological replicates of wild type and ced-9ts strains and 1 of 2 biological replicates of the ifg-1::mos strain. Changes efficiency of ced-9 mRNA translation was determined in ifg-1::mos (A) and ced-9ts (B) strains in comparison to the wild type strain. RNA distributions quantified by qPCR indicating changes in translational efficiency of hif-1 in the ifg-1::mos (C) and ced-9ts (D) strains in comparison to the wild type strains. qPCR quantified RNA distributions indicating changes in the translational efficiency of cep-1 in the ifg-1::mos (E) and ced-9ts (F) strains in comparison to the wild type strain. Changes efficiency of total ced-4 mRNA translation were determined in ifg-1::mos (G) and ced-9ts (H) strains in comparison to the wild type strain. All qPCR data was confirmed in biological duplicates.
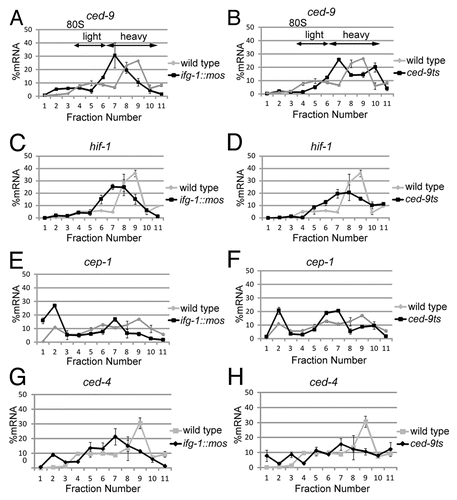
Other stress and apoptotic mRNAs, hif-1 (HIF-1) and cep-1 (p53), have decreased translational efficiency upon depletion of IFG-1 p170
Mammalian hypoxia inducible factor 1 (hif-1) is a transcription factor that promotes cell survival during hypoxia.Citation33 HIF-1 mRNA is translated efficiently under cap-independent conditions during hypoxia in mammalian cell lines.Citation34 p53 mRNA, encoding a key regulator of DNA damage-induced apoptosis, is translated via an IRES.Citation35 We investigated whether C. elegans hif-1 (HIF-1) and cep-1 (p53) mRNAs are also translated more efficiently in ifg-1::mos and ced-9ts worms. Contrary to expectations, decreases in p170 due to the ifg-1 splicing defect ( and ) or its cleavage during apoptosis ( and ) lead to a decrease in hif-1 and cep-1 mRNA translational efficiency. In both cases the hif-1 and cep-1 mRNAs move from very heavy polysomal loading to medium-sized polysomes or, in the case of cep-1 mRNA off polysomes entirely. This decreased efficiency indicates that hif-1 and cep-1 mRNAs require the cap-associated IFG-1 p170 for efficient translation. Thus, despite the importance of HIF-1 and p53 mRNAs during stress conditions in mammalian cell lines, these mRNA do not have a selective translational advantage during stress and/or apoptosis in C. elegans.
ced-4 (Apaf-1) mRNA translation is not induced by IFG-1 p170 depletion
An IRES in the 5′ UTR of mammalian Apaf-1 allows for its synthesis after the induction of cap-independent translation.Citation22 We previously observed that RNAi knockdown of IFG-1 p170 not only increased the number of germ cell corpses, but also induced the appearance of punctate CED-4 (Apaf-1) structures typical of apoptosome formation in dying oocytes.Citation16 Together these data lead us to investigate whether C. elegans ced-4 mRNA also encodes an IRES promoting its cap-independent translation. Due to its integral role in the apoptosome and potential IRES characteristics in its 5′ UTR (see ), we expected that the translational efficiency of ced-4 mRNA to increase after IFG-1 p170 depletion. Contrary to our predictions, ced-4 mRNA underwent a significant decrease in translational efficiency in both the ifg-1::mos () and ced-9ts () strains. Not only does ced-4 message shift to regions in the gradient containing fewer ribosomes, a portion of the mRNA leaves the polysomal region entirely. Thus, the induction of apoptosis and cap-independent conditions inhibit translation initiation of ced-4 mRNA. As this result was inconsistent with the IRES-mediated translation of mammalian Apaf-1, it was necessary to carefully characterize the 5′ structure of ced-4 mRNA to see if such sequences were present in C. elegans mRNA.
Figure 6.ced-4 mRNA structure and mode of translation differs from that of Apaf-1. (A) Diagram comparing C. elegans ced-4 and mammalian Apaf-1 mRNAs. ced-4 is the second gene in an operon with a canonical trans-splice splice site adjacent to its translation start site leaving no encoded 5′ UTR. However, the intergenic region upstream of this splice site has sequence typical of an IRES, such as polypyrimidine tracts and multiple AUG start sites. The arrows indicates the RPA probes extending upstream from exon 1 into the intergenic region. The probes were used to attempt to detect extended ced-4 message variants. (B) RNase Protection Assays were conducted by hybridizing the 5′ probe to total mRNA in wild type, ced-9ts, ifg-1::mos and wild type heat shocked strains. The arrows indicate the SL2 trans-spliced message variant. A band consistent with extended ced-4 mRNA that could not be confirmed with longer ced-4 mRNA probe or other characterization methods (*). (C) RNase Protection Assay using a probe spanning the entire intergenic region. Arrows indicate the detected ced-4 5′ end points. The 3′ UTR of the upstream gene was also protected as indicated by the specified arrows. Control RNase Protection Assay detecting act-1 mRNA is shown to demonstrate RNA quality. (D) Verification of ced-4 mRNA endpoints by circularized RT-PCR. Total mRNA was isolated from C. elegans wild type worms. These mRNAs were decapped using TAP to allow T4 ligation of the 5′ end and the poly(A) tail of mRNAs. cDNA across the junction was then synthesized. An additional nested PCR was performed and the resulting cDNA amplifications were subcloned and sequenced. Agarose gel electrophoresis of nested PCR across the ligated junction is shown. Extended mRNA variants differed due to poly(A) tail size and alternative 3′ end points.
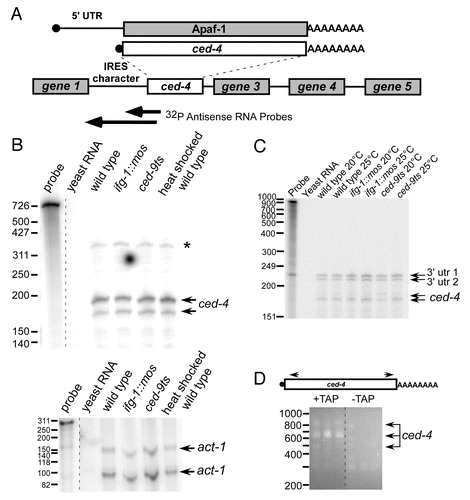
ced-4 mRNA lacks the extended 5′ UTR containing an IRES that is present in Apaf-1 mRNA
We hypothesized that some or all of the conserved intergenic region upstream of the ced-4 translation start site could be present in mature ced-4 mRNA. This region contains sequences characteristic of a putative IRES () that may allow the mRNA to be translated after the knockdown of cap-associated IFG-1 p170. To detect 5′ extended ced-4 mRNA variants, we again performed RNase protection assays (RPAs) on RNA isolated from wild type, ifg-1::mos, ced-9ts and wild type heat shocked () worms using a radiolabeled probe that extended from exon 1 upstream into the intergenic region. Two protected fragment indicative of standard SL2 trans-spliced mRNA that lacked putative IRES sequence were detected in all RNA populations. We did observe a larger band consistent with a longer mRNA variant (*). However, in attempts to confirm this mRNA variant using an RNA probe that extended through the entire intergenic region, we never observed a longer RNA variant (). Additionally, the putative extended mRNA was never detected in polysomal or poly(A) RNA (data not shown). Although two protected fragments were found at 220 and 234 nucleotides with the intergenic sequence probe, these were attributed to the protected 3′ UTR of the upstream gene 1 and are not detected by the shorter probe. As final confirmation of the absence of 5′ extended (IRES) ced-4 mRNA forms, we performed RT-PCR on circularized, end-ligated mRNA using diverging primers in exon 8 and exon 1 (). This procedure amplified the ligated junction between the de-capped 5′ end and poly(A) tail. Only clones beginning with SL2 splice leader sequence adjacent to the start codon were observed. No sequences indicative of a 5′ extended ced-4 mRNA variant were ever found. Therefore, ced-4 mRNA does not encode a 5′ IRES as does its mammalian counterpart. Two 3′ UTR variants for ced-4 mRNA were characterized, including 219 and 257 nt past the termination codon, suggesting alternate polyadenylation sites. The lack of a ced-4 IRES suggests translational regulation of Apaf-1 mRNA differs during nematode apoptosis compared with stressed mammalian cells.
Discussion
Normal cells and tissues utilize both cap-dependent and cap-independent mechanisms of translation initiation to recruit cellular mRNAs.Citation36-Citation38 The relative amount of the cap-independent contribution varies in quiescent, dividing and differentiating cells. It varies as well in response to stress, apoptosis and viral infection.Citation16,Citation39 The contribution is usually dictated by isoforms of eIF4G and their proteolytic cleavage.Citation16,Citation39 In this study we demonstrated a physiological link between levels of cap-associated eIF4G (IFG-1 p170) and translational control of cell death and chaperone mRNAs in whole worms when responding to apoptosis and translational stress. These studies demonstrate that cap-independent translation can play a role in the natural apoptotic cell fate decision in healthy, whole organisms and their natively differentiating cell lineage (e.g., germ cells). Genetic depletion of a key translation initiation factor (IFG-1 p170), or its cleavage during ced-9ts-induced apoptosis, caused a subset of mRNAs to increase in translational efficiency. This illustrates that these mRNAs may utilize cleaved IFG-1 p170 or constitutively non-cap associated p130 for their initiation. Conditions that produced enhanced cap-independent initiation favored more efficient translation of C. elegans BiP (hsp-3) mRNA in vivo. As demonstrated, these conditions may be either genetic depletion of IFG-1 p170 (eIF4G) or CED-9 (Bcl-2) inactivation that leads to IFG-1 p170 cleavage by CED-3 (caspase). Ultimately, the translational mechanism may be identical. These data are consistent with translational enhancement of the mammalian BiP mRNA after the induction of cap-independent protein synthesis.Citation40 The mammalian BiP is translated cap-independently via an IRES sequence in its 5′ UTR.Citation41 Although Li et al. demonstrated enhanced expression of a downstream cistron from the hsp-3 putative 5′ UTR in a bicistronic transgenic reporter, our RNase protection mapping of hsp-3 mRNA 5′ ends indicates that all of the detectable hsp-3 transcripts have short (~25 nt) 5′ UTRs that lack the purported IRES.Citation27 The sequence used by Li et al. to drive bicistronic expression also contained an intron sequence that is spliced out of the single known, extended hsp-3 expressed sequence tag.Citation42 The hsp-3 upstream intron has been reported to contain enhancer elements and a TATA box that appear to promote transcription initiation from the start site that we have mapped.Citation30,Citation29 Our data indicate that short hsp-3 mRNAs translate efficiently cap-independently and their cap-independent translation is not dependent on the purported IRES. There are numerous precedents showing heat shock protein mRNAs that lack IRESes still translate by cap-independent mechanisms in stressed mammalian and plant cells, and in virus infected cells.Citation43-Citation46 C. elegans hsp-3 mRNA is clearly one of these cap- and IRES-independent mRNAs. The translational efficiency of ced-9 (Bcl-2) mRNA also increased after CED-9 inactivation. The disruption of Bcl-2 function in living worms allowed us to look at physiologically induced change in IFG-1 isoform representation leading to enhanced ced-9 mRNA translation (producing an inactive protein). These experiments illustrate the importance of eIF4G (IFG-1 p170 and p130) isoform representation in selecting mRNAs for translation in response to stressors to the organism.
It is notable that the differences in mRNA translational efficiency were observed as changes in polysomal mRNA distribution in total worm lysates. Certainly apoptotic or stressor stimuli in individual cell types or tissues would lead to more robust recruitment of unique mRNAs for translation than we document here. Most likely the changes we observed in translational efficiency were muted due to the large number of somatic and germ cells not undergoing apoptosis. We expect, then, that changes in translational efficiency within individual cells result in large changes in protein synthesis that may determine their differentiated vs. apoptotic state. Clearly, the changes in translation initiation were robust enough to enhance germ cell apoptosis and decrease fecundity. The number of germ cell corpses increased to a level equivalent to that observed when the apoptotic pathway was directly affected. As expected, increased apoptosis decreased the fertility of ifg-1::mos adult hermaphrodites. We also observed that constricting cap-dependent translation resulted in a high level of embryonic lethality and arrested larval development. The multiple developmental arrests support our previous findings on the importance of cap-dependent protein synthesis to normal worm development.Citation15,Citation47
Several putative cap-independent IRES mRNAs did not respond similarly to cap-independent conditions in whole worms. Although HIF-1, Apaf-1 and p53 mRNAs are reported to translate cap-independently in mammalian cell culture via IRES sequences, we did not observe similar translational enhancement in C. elegans. Instead, C. elegans homologs of these mRNAs translated less efficiently when cap-associated IFG-1 p170 was depleted. hif-1 (HIF-1), cep-1 (p53), and ced-4 (Apaf-1) mRNAs were markedly absent from heavy polysomes suggesting they were at a disadvantage for initiation (). In several cases the C. elegans homolog mRNA differs significantly from the mammalian counterpart. In particular, unlike its mammalian homolog Apaf-1, ced-4 was shown by mapping to lack a 5′ IRES sequence and translated poorly in both ifg-1::mos and ced-9ts genetic backgrounds.Citation22 We previously observed that the RNAi knockdown of IFG-1 p170 led to the accumulation of punctate CED-4 structures typical of apoptosomes in apoptosing germ cells.Citation16 The current study suggests their appearance of CED-4 structures was probably not due to new synthesis, but may have resulted from enhanced protein stability and the accumulation of CED-4 into easily detected foci on apoptosomes. More importantly, our data point out clear differences between protein synthetic regulation of stress and cell death proteins such as Apaf-1, p53 and HIF-1 in whole animals vs. cells in culture. Such observations suggest that both organism and physiological context may greatly alter expected protein synthetic regulation
Figure 7. Model for the induction of cap-independent translation of stress-related mRNAs during apoptosis. A. Model comparing the translational efficiencies of C. elegans and mammalian stress related mRNAs. Wide width arrows indicate the increased translational efficiency of C. elegans hsp-3 and ced-9 mRNAs and human BiP, Bcl-2, Apaf-1, Hif-1 and p53. The decreased translational efficiency of other C. elegans mRNAs, ced-4, hif-1 and cep-1, is shown by an “x.” * indicates the genetic manipulation of the ifg-1::mos strain. B. The induction of apoptosis in humans leads to the activation of caspases and cleavage of the cap-dependent translation initiation factor eIF4GI and constitutively cap-independent translation initiation factor p97. This cleavage leads to the induction of cap-independent protein synthesis. As a result, the translational efficiency is increased for pro- and anti-apoptotic messages such as pro-apoptotic Apaf-1 and recovery mRNAs Bcl-2 and BiP. The translation of Bcl-2 and BiP promotes periods of cellular recovery from stress. Subsequent translation of Apaf-1 leads to cellular suicide. In the C. elegans gonad, apoptosis activated caspase, CED-3 leads to C. elegans eIF4GI (IFG-1 p170) and p97 (IFG-1 p130) cleavage. This cleavage leads to an increase in p130-driven translation of stress-related mRNAs hsp-3 and ced-9. * indicates that genetic manipulation in the ifg-1::mos and ced-9ts strains leads to the induction of cap-independent protein synthesis. # indicates a proapoptotic function for CED-9.Citation32
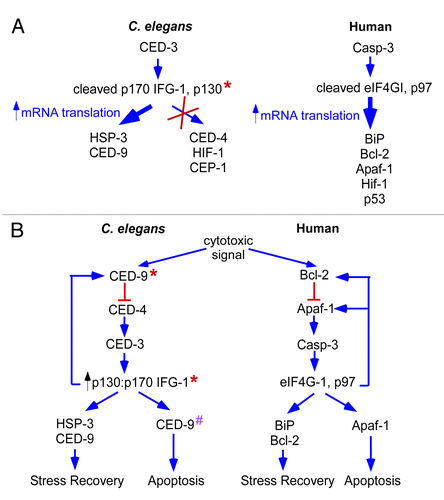
We propose a model that relates protein synthesis to the apoptotic pathway in normal development and physiological stress (). In this model, the translation machinery is itself responsible for the selection of mRNAs during times of stress. This allows a cell to initially attempt recovery before committing itself to cellular suicide. Cell death signals initiates apoptosis by preventing CED-9 (Bcl-2) from inhibiting the activity of CED-4 (Apaf-1).Citation26,Citation48,Citation49 In our experiments the genetic loss of CED-9 (Bcl-2) function allows precocious activation of CED-4 (Apaf-1). CED-4 binds procaspase CED-3 forming the apoptosomeCitation50,Citation51 Apoptosome formation initiates autocatalysis and activation of CED-3.Citation51,Citation52 Active CED-3 then cleaves IFG-1(eIF4G) separating the PABP and eIF4E-binding domains from the region that associates with the ribosome.Citation15 This prevents utilization of the mRNA cap or poly(A) for mRNA recruitment to the ribosome. Instead, it promotes cap-independent mRNA initiation through the ribosome-binding portion of IFG-1 p130 or the p130-like cleavage product of p170.Citation15 Cap-independent translation of mRNAs (either containing or lacking IRESes) allows for the expression first of proteins involved in attempted recovery, then subsequently those that ensure the apoptotic fate. One such non-IRES mRNA translationally enhanced during the initial phase is hsp-3 (BiP). This chaperone protein is thought to promote protein refolding under stress conditions.Citation20 CED-9 (Bcl-2) synthesis is also enhanced and may also be involved in the recovery phase. Since CED-9 is an integral member of the apoptotic pathway, the induction of cap-independent translation may result in a feedback loop that prevents further apoptosis. Interestingly, some cap-independent products switch roles to promote apoptosis at later times if recovery has not been accomplished. Genetic evidence shows that CED-9 may be one of these proteins with an initial role in cellular survival followed by a later role in cell death.Citation32 In mammalian cells, a similar initial attempt at recovery involves the sequential use of first anti-apoptotic and then pro-apoptotic IRESes for translation control during progressive stress conditions.Citation18 In both cases, regulated cap-independent translation mediates the switch.
We are discovering that the interplay of cap-dependent and -independent translation during development may provide a means to selectively enhance translational efficiency of mRNAs that influence cell fate decisions. Germ cells in particular must rely on translational regulation of stored maternal mRNAs for their maturation. Alternatively, germ cells undergo apoptosis inducing translation initiation factor eIF4G (IFG-1) cleavage. At the threshold of that decision, cap-independent synthesis of anti-apoptotic CED-9 and HSP-3 may suppress germ cell apoptosis and allow most cells to mature into oocytes.
Methods
Strains
The wild type-type strain used in experiments was MD701, {bcIs39 V [Plim-7 ced-1::gfp and lin-15(+)]} and was obtained from Barbara Conradt. The CED-1::GFP reporter was used to assay germ cell apoptosis as previously described.Citation23 The original ifg-1::mos strain {ifg-1(cxP9279)/+, with a mos-1 transposon insertion} was made by Laurent Segalat, (CNRS) using mobilization of Mos-1 (mariner-1, Drosophila) by SP:Mos transposase.Citation53,Citation54 This line contained a stable insertion of Mos-1 transposon into ifg-1 (M110.4) at 17,266 nt (intron 5). ifg-1::mos/+ males were then crossed into a strain carrying the mT1 balancer. Progeny were outcrossed five time to wild type (N2), then self crossed to homozygous ifg-1::mos allele in the strain, KX34. KX34 was crossed with the MD701 strain to produce an ifg-1::mos strain easily scored for germ line apoptosis (KX54). The ced-9ts strain {ced-9 (n1653) mab-5 (mu114)} was obtained from the Caenorhabditis Genetics center and was also crossed with the MD701 strain creating the resulting KX110 strain. All strains were grown on normal growth medium (NGM) plates with E. coli strain OP50 at 20 °C.Citation55 Strains were shifted to 25 °C for 24 h prior to harvesting and liquid nitrogen pelleting.
RNA isolation, Northern Blotting, RT-PCR and cRT-PCR
Total RNA was extracted and purified as previously described using the Trizol (Invitrogen) method.Citation16 Northern Blotting was performed using an antisense ifg-1 riboprobe complementary to the 5′ half of the ifg-1 cDNA or to the gpd-3 cDNA. Blot imaging was done on a Typhoon 9410 (GE Health Care) and quantification of mRNAs was done with ImageQuant TL 2003 software (GE Health Care). ifg-1 mRNA signal was normalized to gpd-3 signal to account for total RNA loading. Sequences for the ifg-1 mRNAs containing Mos insertions were amplified using SuperScript II (Invitrogen) with internal and external primers (5′-ACCAAACTGGGCAAACAAAG-3′, 5′-GCTCAATTCGCGCCAAACTATG-3′, 5′-ACATCGACTGCTCGACCGAA-3′, 5′-GCCGTCGCTAGCTACACATTT-3′, and 5′-GCGCATCGTTGGAGGATACA-3′) and subcloned into pCRII using Topo cloning (Invitrogen). cRT-PCR was performed as previously describedCitation15 using primary ced-4 primers, forward 5′-GAAGACTTCCCAAAGTTCATGCA-3′ and reverse 5′-CTTCGAGGAAATCAGCAAGG-3′ and nested ced-4 primers, forward 5′-GCACCAGAAATTCTATGACTCCCT-3′ and reverse 5′-TGGTTCAAAGTCGTGGATGA-3′.
Sucrose gradient fractionation of polysomes and qPCR
Sucrose gradient fractionation was performed as detailed previously with the exception that gradient fractions were isolated in 4 volumes Trizol.Citation47 RNA was prepared from half of the 1 mL fractions, storing the duplicate halves at -80 °C. Verification of polysome release was accomplished in EDTA-treated duplicate gradients in which EDTA was added to the lysates immediately before overlaying on the gradients and cycloheximide was excluded (data not shown). 0.25 µg of RNA from each fraction or 1 µg total RNA was reverse transcribed into cDNA using the iScript cDNA Synthesis Kit (BioRad). Real-time PCR was performed in triplicate using the iQ SYBR Green Supermix (BioRad) in an iCycler iQ5 Real-time PCR machine according the manufacture’s protocol. The specific primer sequences used were: gpd-3 (forward 5′-GATCTCAGCTGGGTCTCTT-3′, reverse 5′-TCCAGTACGATTCCACTCAC-3′); hsp-3 (forward 5′-GAGGATGACAAGAAGGTCAA-3′, reverse 5′-TCTTCAATCTGGTTTTTCAGG-3′); ced-4 (forward 5′- GACAAATGCTCGATAGGAAA-3′, reverse 5′-GCCGTGTAGAAACAGAAAAA-3′); hif-1 (forward 5′- GACATCATACGGACGAAGAG -3′, reverse 5′- CAAGTGGCGAGTAGTTGG -3′); cep-1 (forward 5′- TTCGTTTAGAACGCTCACTC-3′, reverse 5′- GTATCTGGGAACTTTTGCTTC -3′); hsp-4 (forward 5′- TGCCTCTACTGAAGAAAACAA -3′, reverse 5′- CGAGTAAAGTTTGGAGACGA-3′); ced-9 (forward 5′- CGTTTGATAGGTCTAATCTCGT -3′, reverse 5′- ATTGTGTTCCTTCCAGTTGT -3′). The qPCR contents were normalized to total RNA content to accurately report mRNA translational efficiencies.
Western Blotting and RNase protection assays
Western blots were performed using the central IFG-1 antibody that detects both the p130 and p170 protein isoforms as previously described.Citation16 Actin was detected using an anti-chicken actin antibody (Sigma). Blot imaging was done using ECL+ Detection System (GE Health Care) on a Typhoon 9410 and protein quantification was done with ImageQuant TL 2003 software (GE Health Care). IFG-1 p130 and p170 signals were normalized to actin to account for total protein loading. For RNase Protection assay of ced-4 mRNA, an antisense RNA probe uniformly labeled with [32P]UTP was used extending from 156 bp into exon 1 and containing 186 bp of the 5′ UTR. A second ced-4 mRNA probe extended from 176 bp into exon 1 and contained 644bp of the intergenic region and upstream gene. The hsp-3 mRNA probe extended from72 bp into exon 1 of the hsp-3 gene to the end of the longest 5′ UTR detected by ESTs (268 bp upstream of the ATG). This probe was designed using the hsp-3 EST and thus did not contain the intron reported in the 5′ UTR. The assay was performed as previously described with the exception that the products were resolved on an 8% polyacrylamide/urea/TBE gel.Citation16
Microscopy and determination of the extent of apoptosis
Microscopy was performed on a Zeiss Axiovert 200M as previously described on unfixed worms from strains KX110 {ced-9ts (n1653), CED-1::GFP}, MD701 (CED-1::GFP), and KX54 (ifg-1::mos, CED-1::GFP) after a temperature shift of L3 worms to 25 °C for 0, 12, 24, 36, and 48 h.Citation15 Worms frozen for RNA isolation were grown on mixed population egg plates that were shifted to 25 °C 24 h prior to liquid nitrogen pelleting. All worms visualized for microscopy were immobilized with 0.5 µM levamisole in M9 for time course experiments and 30 nM Sodium Azide in M9 for all other experiments. Fertility assays on wild type and ifg-1::mos strains were conducted at 25 °C. The visual morphology of the worms was determined at 24-h intervals.
Cloning and sequencing
Cloning of PCR products was performed using TOPO TA cloning kits (Invitrogen) with cloning vector pCRII according to the manufacturer’s instructions. Plasmids were grown in TOP 10 cells. Sequencing was completed by the DNA sequencing facility at Iowa State University.
| Abbreviations: | ||
| IRES | = | internal ribosome entry sites |
| BiP/HSP-3/4 | = | binding immunoglobulin protein/heat shock protein-3/4 |
| HIF-1 | = | hypoxia-inducible factor 1 |
| p53/CEP-1 | = | tumor protein 53/C. elegans p53-like protein |
| Bcl-2/CED-9 | = | B-cell lymphoma 2/cell death abnormality 9 |
| Apaf-1/CED-4 | = | apoptotic protease activating factor 1/cell death abnormality 4 |
| IFG-1 | = | initiation factor 4G gene 1 of C. elegans |
| eIF4GI | = | eukaryotic initiation factor 4G gene 1 |
| p97 | = | human 97 kDa eIF4G-homologous protein |
| eIF4E | = | eukaryotic initiation factor 4E |
| PABP | = | poly(A) binding protein |
| c-Myc | = | cellular myelocytomatosis |
| CED-1 | = | cell death abnormality 1 |
| GFP | = | green fluorescent protein |
| RNAi | = | RNA interference |
| L3 | = | larval 3 growth stage |
| UTR | = | untranslated region |
| qPCR | = | quantitative reverse transcriptase-polymerase chain reaction |
| GAPDH/GPD-3 | = | glyceraldehyde 3-phosphate dehydrogenase |
| ITAFs | = | IRES-interacting trans-acting factors |
| SL2 trans-splicing | = | splice leader 2 trans-splicing |
| RPA | = | RNase Protection Assay |
| 4E-BP | = | eIF4E binding protein |
| NGM | = | normal growth medium |
| RT-PCR | = | reverse transcriptase-polymerase chain reaction |
| cRT-PCR | = | circularized reverse transcriptase-polymerase chain reaction |
| TAP | = | tobacco acid pyrophosphatase |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank the Caenorhabditis Genetics Center for the ced-9ts (n1653) strain. We also thank Dr. Yuji Kohara (National Institute of Genetics, Japan) for several hsp-3 and hsp-4 cDNAs, and Dr. Vince Contreras (National Institutes of Health) for improving this manuscript and many useful discussions. This work was supported by grant MCB0842475 from the National Science Foundation.
References
- Kimble J, Crittenden SL. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annu Rev Cell Dev Biol 2007; 23:405 - 33; http://dx.doi.org/10.1146/annurev.cellbio.23.090506.123326; PMID: 17506698
- Gumienny TL, Lambie E, Hartwieg E, Horvitz HR, Hengartner MO. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development 1999; 126:1011 - 22; PMID: 9927601
- Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J 1998; 17:7480 - 9; http://dx.doi.org/10.1093/emboj/17.24.7480; PMID: 9857202
- Lamphear BJ, Kirchweger R, Skern T, Rhoads RE. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J Biol Chem 1995; 270:21975 - 83; http://dx.doi.org/10.1074/jbc.270.37.21975; PMID: 7665619
- Gradi A, Imataka H, Svitkin YV, Rom E, Raught B, Morino S, Sonenberg N. A novel functional human eukaryotic translation initiation factor 4G. Mol Cell Biol 1998; 18:334 - 42; PMID: 9418880
- Imataka H, Olsen HS, Sonenberg N. A new translational regulator with homology to eukaryotic translation initiation factor 4G. EMBO J 1997; 16:817 - 25; http://dx.doi.org/10.1093/emboj/16.4.817; PMID: 9049310
- Morino S, Imataka H, Svitkin YV, Pestova TV, Sonenberg N. Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol Cell Biol 2000; 20:468 - 77; http://dx.doi.org/10.1128/MCB.20.2.468-477.2000; PMID: 10611225
- Keiper BD, Gan W, Rhoads RE. Protein synthesis initiation factor 4G. Int J Biochem Cell Biol 1999; 31:37 - 41; http://dx.doi.org/10.1016/S1357-2725(98)00130-7; PMID: 10216942
- Bushell M, McKendrick L, Jänicke RU, Clemens MJ, Morley SJ. Caspase-3 is necessary and sufficient for cleavage of protein synthesis eukaryotic initiation factor 4G during apoptosis. FEBS Lett 1999; 451:332 - 6; http://dx.doi.org/10.1016/S0014-5793(99)00614-6; PMID: 10371215
- Pestova TV, Shatsky IN, Hellen CU. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol 1996; 16:6870 - 8; PMID: 8943342
- Marissen WE, Lloyd RE. Eukaryotic translation initiation factor 4G is targeted for proteolytic cleavage by caspase 3 during inhibition of translation in apoptotic cells. Mol Cell Biol 1998; 18:7565 - 74; PMID: 9819442
- Henis-Korenblit S, Strumpf NL, Goldstaub D, Kimchi A. A novel form of DAP5 protein accumulates in apoptotic cells as a result of caspase cleavage and internal ribosome entry site-mediated translation. Mol Cell Biol 2000; 20:496 - 506; http://dx.doi.org/10.1128/MCB.20.2.496-506.2000; PMID: 10611228
- Marissen WE, Gradi A, Sonenberg N, Lloyd RE. Cleavage of eukaryotic translation initiation factor 4GII correlates with translation inhibition during apoptosis. Cell Death Differ 2000; 7:1234 - 43; http://dx.doi.org/10.1038/sj.cdd.4400750; PMID: 11175261
- Nevins TA, Harder ZM, Korneluk RG, Holcík M. Distinct regulation of internal ribosome entry site-mediated translation following cellular stress is mediated by apoptotic fragments of eIF4G translation initiation factor family members eIF4GI and p97/DAP5/NAT1. J Biol Chem 2003; 278:3572 - 9; http://dx.doi.org/10.1074/jbc.M206781200; PMID: 12458215
- Contreras V, Friday AJ, Morrison JK, Hao E, Keiper BD. Cap-independent translation promotes C. elegans germ cell apoptosis through Apaf-1/CED-4 in a caspase-dependent mechanism. PLoS One 2011; 6:e24444; http://dx.doi.org/10.1371/journal.pone.0024444; PMID: 21909434
- Contreras V, Richardson MA, Hao E, Keiper BD. Depletion of the cap-associated isoform of translation factor eIF4G induces germline apoptosis in C. elegans. Cell Death Differ 2008; 15:1232 - 42; http://dx.doi.org/10.1038/cdd.2008.46; PMID: 18451872
- De Benedetti A, Graff JR. eIF-4E expression and its role in malignancies and metastases. Oncogene 2004; 23:3189 - 99; http://dx.doi.org/10.1038/sj.onc.1207545; PMID: 15094768
- Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol 2005; 6:318 - 27; http://dx.doi.org/10.1038/nrm1618; PMID: 15803138
- Yang Q, Sarnow P. Location of the internal ribosome entry site in the 5′ non-coding region of the immunoglobulin heavy-chain binding protein (BiP) mRNA: evidence for specific RNA-protein interactions. Nucleic Acids Res 1997; 25:2800 - 7; http://dx.doi.org/10.1093/nar/25.14.2800; PMID: 9207027
- Lee AS. The ER chaperone and signaling regulator GRP78/BiP as a monitor of endoplasmic reticulum stress. Methods 2005; 35:373 - 81; http://dx.doi.org/10.1016/j.ymeth.2004.10.010; PMID: 15804610
- Sherrill KW, Byrd MP, Van Eden ME, Lloyd RE. BCL-2 translation is mediated via internal ribosome entry during cell stress. J Biol Chem 2004; 279:29066 - 74; http://dx.doi.org/10.1074/jbc.M402727200; PMID: 15123638
- Coldwell MJ, Mitchell SA, Stoneley M, MacFarlane M, Willis AE. Initiation of Apaf-1 translation by internal ribosome entry. Oncogene 2000; 19:899 - 905; http://dx.doi.org/10.1038/sj.onc.1203407; PMID: 10702798
- Zhou Z, Hartwieg E, Horvitz HR. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell 2001; 104:43 - 56; http://dx.doi.org/10.1016/S0092-8674(01)00190-8; PMID: 11163239
- Hengartner MO, Ellis RE, Horvitz HR. Caenorhabditis elegans gene ced-9 protects cells from programmed cell death. Nature 1992; 356:494 - 9; http://dx.doi.org/10.1038/356494a0; PMID: 1560823
- Chinnaiyan AM, O’Rourke K, Lane BR, Dixit VM. Interaction of CED-4 with CED-3 and CED-9: a molecular framework for cell death. Science 1997; 275:1122 - 6; http://dx.doi.org/10.1126/science.275.5303.1122; PMID: 9027312
- Pourkarimi E, Greiss S, Gartner A. Evidence that CED-9/Bcl2 and CED-4/Apaf-1 localization is not consistent with the current model for C. elegans apoptosis induction. Cell Death Differ 2012; 19:406 - 15; http://dx.doi.org/10.1038/cdd.2011.104; PMID: 21886181
- Li D, Wang M. Construction of a bicistronic vector for the co-expression of two genes in Caenorhabditis elegans using a newly identified IRES. Biotechniques 2012; 52:173 - 6; PMID: 22401550
- Cho S, Park SM, Kim TD, Kim JH, Kim KT, Jang SK. BiP internal ribosomal entry site activity is controlled by heat-induced interaction of NSAP1. Mol Cell Biol 2007; 27:368 - 83; http://dx.doi.org/10.1128/MCB.00814-06; PMID: 17074807
- Heschl MF, Baillie DL. The HSP70 multigene family of Caenorhabditis elegans. Comp Biochem Physiol B 1990; 96:633 - 7; http://dx.doi.org/10.1016/0305-0491(90)90206-9; PMID: 2225768
- Heschl MF, Baillie DL. Characterization of the hsp70 multigene family of Caenorhabditis elegans. DNA 1989; 8:233 - 43; http://dx.doi.org/10.1089/dna.1.1989.8.233; PMID: 2766926
- Sherrill KW, Byrd MP, Van Eden ME, Lloyd RE. BCL-2 translation is mediated via internal ribosome entry during cell stress. J Biol Chem 2004; 279:29066 - 74; http://dx.doi.org/10.1074/jbc.M402727200; PMID: 15123638
- Galvin BD, Denning DP, Horvitz HR. SPK-1, an SR protein kinase, inhibits programmed cell death in Caenorhabditis elegans. Proc Natl Acad Sci U S A 2011; 108:1998 - 2003; http://dx.doi.org/10.1073/pnas.1018805108; PMID: 21245325
- Keith B, Simon MC. Hypoxia-inducible factors, stem cells, and cancer. Cell 2007; 129:465 - 72; http://dx.doi.org/10.1016/j.cell.2007.04.019; PMID: 17482542
- Lang KJ, Kappel A, Goodall GJ. Hypoxia-inducible factor-1alpha mRNA contains an internal ribosome entry site that allows efficient translation during normoxia and hypoxia. Mol Biol Cell 2002; 13:1792 - 801; http://dx.doi.org/10.1091/mbc.02-02-0017; PMID: 12006670
- Weingarten-Gabbay S, Khan D, Liberman N, Yoffe Y, Bialik S, Das S, Oren M, Kimchi A. The translation initiation factor DAP5 promotes IRES-driven translation of p53 mRNA. Oncogene 2013; PMID: 23318444
- Gilbert WV. Alternative ways to think about cellular internal ribosome entry. J Biol Chem 2010; 285:29033 - 8; http://dx.doi.org/10.1074/jbc.R110.150532; PMID: 20576611
- Malys N, McCarthy JE. Translation initiation: variations in the mechanism can be anticipated. Cell Mol Life Sci 2011; 68:991 - 1003; http://dx.doi.org/10.1007/s00018-010-0588-z; PMID: 21076851
- Komar AA, Mazumder B, Merrick WC. A new framework for understanding IRES-mediated translation. Gene 2012; 502:75 - 86; http://dx.doi.org/10.1016/j.gene.2012.04.039; PMID: 22555019
- Keiper BD, Rhoads RE. Cap-independent translation initiation in Xenopus oocytes. Nucleic Acids Res 1997; 25:395 - 402; http://dx.doi.org/10.1093/nar/25.2.395; PMID: 9016570
- Buchkovich NJ, Yu Y, Pierciey FJ Jr., Alwine JC. Human cytomegalovirus induces the endoplasmic reticulum chaperone BiP through increased transcription and activation of translation by using the BiP internal ribosome entry site. J Virol 2010; 84:11479 - 86; http://dx.doi.org/10.1128/JVI.01330-10; PMID: 20739513
- Koong AC, Auger EA, Chen EY, Giaccia AJ. The regulation of GRP78 and messenger RNA levels by hypoxia is modulated by protein kinase C activators and inhibitors. Radiat Res 1994; 138:Suppl S60 - 3; http://dx.doi.org/10.2307/3578763; PMID: 8146329
- WormBase. hsp-3. In: R. Durbin JS, L. Stein and P. Sternberg, ed. WormBase: C elegans online database, 2007.
- Dinkova TD, Keiper BD, Korneeva NL, Aamodt EJ, Rhoads RE. Translation of a small subset of Caenorhabditis elegans mRNAs is dependent on a specific eukaryotic translation initiation factor 4E isoform. Mol Cell Biol 2005; 25:100 - 13; http://dx.doi.org/10.1128/MCB.25.1.100-113.2005; PMID: 15601834
- Rhoads RE, Lamphear BJ. Cap-independent translation of heat shock messenger RNAs. Curr Top Microbiol Immunol 1995; 203:131 - 53; http://dx.doi.org/10.1007/978-3-642-79663-0_7; PMID: 7555088
- Joshi-Barve S, De Benedetti A, Rhoads RE. Preferential translation of heat shock mRNAs in HeLa cells deficient in protein synthesis initiation factors eIF-4E and eIF-4 gamma. J Biol Chem 1992; 267:21038 - 43; PMID: 1400417
- Schneider RJ. Cap-independent translation in adenovirus infected cells. Curr Top Microbiol Immunol 1995; 203:117 - 29; http://dx.doi.org/10.1007/978-3-642-79663-0_6; PMID: 7555087
- Henderson MA, Cronland E, Dunkelbarger S, Contreras V, Strome S, Keiper BD. A germline-specific isoform of eIF4E (IFE-1) is required for efficient translation of stored mRNAs and maturation of both oocytes and sperm. J Cell Sci 2009; 122:1529 - 39; http://dx.doi.org/10.1242/jcs.046771; PMID: 19383718
- Hengartner MO, Horvitz HR. C. elegans cell survival gene ced-9 encodes a functional homolog of the mammalian proto-oncogene bcl-2. Cell 1994; 76:665 - 76; http://dx.doi.org/10.1016/0092-8674(94)90506-1; PMID: 7907274
- Zou H, Henzel WJ, Liu X, Lutschg A, Wang X. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell 1997; 90:405 - 13; http://dx.doi.org/10.1016/S0092-8674(00)80501-2; PMID: 9267021
- Nicholson DW, Ali A, Thornberry NA, Vaillancourt JP, Ding CK, Gallant M, Gareau Y, Griffin PR, Labelle M, Lazebnik YA, et al. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature 1995; 376:37 - 43; http://dx.doi.org/10.1038/376037a0; PMID: 7596430
- Chinnaiyan AM, Chaudhary D, O’Rourke K, Koonin EV, Dixit VM. Role of CED-4 in the activation of CED-3. Nature 1997; 388:728 - 9; http://dx.doi.org/10.1038/41913; PMID: 9285582
- Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, Wang X. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell 1997; 91:479 - 89; http://dx.doi.org/10.1016/S0092-8674(00)80434-1; PMID: 9390557
- Bessereau JL, Wright A, Williams DC, Schuske K, Davis MW, Jorgensen EM. Mobilization of a Drosophila transposon in the Caenorhabditis elegans germ line. Nature 2001; 413:70 - 4; http://dx.doi.org/10.1038/35092567; PMID: 11544527
- Granger L, Martin E, Ségalat L. Mos as a tool for genome-wide insertional mutagenesis in Caenorhabditis elegans: results of a pilot study. Nucleic Acids Res 2004; 32:e117; http://dx.doi.org/10.1093/nar/gnh111; PMID: 15310838
- Brenner S. The genetics of Caenorhabditis elegans. Genetics 1974; 77:71 - 94; PMID: 4366476
