Abstract
Over the past 20 years, a better understanding of cancer biology, screening for early detection, improved adjuvant treatment, and targeted therapies have decreased the rate of breast cancer deaths. However, resistance to treatment is common, and new approaches are needed. Deregulation of translation initiation is associated with the commencement and progression of cancer. Often, translation initiation factors are overexpressed and the related signaling pathways activated in human tumors. Recently, a significant number of inhibitors that target translation factors and pathways have become available. These inhibitors are being tested alone or in combination with chemotherapeutic agents in clinical trials. The results are varied, and it is not yet clear which drug treatments most effectively inhibit tumor growth. This review highlights the pathways and downstream effects of the activation of translation and discusses targeting the control of translation initiation as a therapeutic approach in cancer, focusing on breast cancer clinical trials.
Introduction
Breast cancer is the second-most frequently diagnosed cancer and is the second-most frequent cause of cancer-related death in women.Citation1 In the past decade, its incidence rates have not decreased. However, since 1989, early detection and better treatment have decreased mortality rates from breast cancer. Current treatment options include surgical removal of the tumor and radiation therapy (depending on surgery type and clinical variables), and with recurrence, chemotherapy in the adjuvant or neoadjuvant setting, endocrine therapy (e.g., estrogen response modifiers, aromatase inhibitors, and ovarian ablation), and recently, targeted therapy.
Regulation of mRNA translation to protein is a major point of control for gene expression. Recently, deregulation of protein synthesis and its involvement in cancer development have become the subject of much discussion.Citation2-Citation6 Cancer cells are at a metabolically demanding state, which requires an increase in protein synthesis. Thus, sustained activation of protein expression may not only initiate but also maintain cancer growth. In addition, to increase overall protein synthesis, deregulation results in the activation of signaling pathways and synthesis of various proteinsCitation3 required for tumorigenesis. These proteins may facilitate cancer progression or help cancer cells develop resistance to treatment. Therefore, alterations in translation may activate major survival pathways. A significant amount of data suggests that translation initiation is an integral part of carcinogenesis, including breast cancer development. This review discusses the rapidly growing field of translation initiation inhibition in breast cancer.
A Summary of Translational Control
mRNA translation to protein can be divided into four stages: initiation, elongation, termination, and ribosome recycling.Citation7 Translation initiation is the limiting step in this process.Citation8 In higher eukaryotes, including humans, most mRNA translation is cap-dependent, relying on the complex of proteins that assemble at the 5′ end of mRNA, which contains the 7-methylguanosine cap (5′ cap) structure.Citation7,Citation9,Citation10 This complex, called the eukaryotic initiation factor (eIF) 4F (eIF4F) complex, consists of the cap-binding protein eIF4E, the scaffold protein eIF4G, and the RNA helicase protein eIF4A. Polyadenylate-binding protein attaches to the 3′ poly(A) tail and to eIF4G and brings the 3′ and 5′ ends of mRNA together.Citation11 This is followed by the binding of eIF4B, which interacts with eIF4F and eIF4A, further stabilizing the complex. Meanwhile, the 40S ribosomal subunit binds to additional initiation factors and a ternary complex (eIF2/GTP/met-tRNA) to form the 43S pre-initiation complex. Interacting with eIF3 and the scaffolding protein eIF4G, the 43S complex binds to the 5′ cap. The cap-initiation complex scans the mRNA in a 5′-to-3′ direction until it encounters a start codon. Recognition of a start codon results in hydrolysis of GTP bound to eIF2 and release of eIFs proteins. Once the 40S subunit is positioned and the 60S subunit recruited, an elongation-competent 80S ribosome is formed, ready to begin protein synthesis.
Major signaling pathways, such as the phosphatidylinositol 3-hydroxy kinase (PI3K)/Akt/mammalian target of rapamycin (mTOR) pathway, also participate in translation initiation. These steps indicate that deregulation of translation initiation can have a key role in cancer development and maintenance. Therefore, targeting the factors and pathways involved in this process with inhibitors may offer novel treatment options for overcoming resistance to chemotherapeutics.
Activation of mitogen-activated protein kinases (MAPKs), PI3K/Akt/mTOR signaling, RAS signaling, and receptor tyrosine kinases, such as human epidermal growth factor receptor 2 (HER2), EGFR, and IGF1R, may contribute to tumor growth and aggressiveness (). These pathways regulate both global translation and the translation of specific mRNAs that take part in cell proliferation and survival. The PI3K/Akt/mTOR pathway is deregulated through several mechanisms, including overexpression or activation of growth factor receptors (including HER2), mutation or amplification of PI3K or of Akt, amplification or overexpression of PDK1, and decreased expression of tumor suppressor phosphatase and tensin homolog.Citation12,Citation13 Akt controls translation initiation mainly through activation of mTOR complex 1 (mTORC1).Citation13 mTORC1 phosphorylates eIF4E-binding proteins (4E-BPs). The 4E-BPs compete with eIF4G for binding to eIF4E, thus preventing eIF4G recruitment. Once hypophosphorylated, 4E-BPs release and activate eIF4E. Increased availability of eIF4E is associated with progression and poor survival in several types of cancer, including breast cancer.Citation14,Citation15 In addition, mTORC1 regulates cell growth through ribosomal proteins S6 kinase 1 (S6K1) and S6 kinase 2. Phosphatase and tensin homolog, tuberous sclerosis 1, and tuberous sclerosis 2 mutations also activate S6K1 and S6 kinase 2. S6K1 phosphorylates eIF4B, which in turn stimulates eIF4A helicase activity.
Figure 1. Regulatory networks involved in translation initiation and points of inhibition. Arrows represent activation, and bars represent inhibition. Small-molecule inhibitors of the signaling nodes are listed below. Abbreviations: 4E-BP, eukaryotic initiation factor 4E–binding protein; AMPK, adenosine monophosphate–activated protein kinase; eEF2, eukaryotic elongation factor 2; eEF2K, eukaryotic elongation factor 2 kinase; eIF2B, eukaryotic initiation factor 2B; eIF4A, eukaryotic initiation factor 4A; eIF4B, eukaryotic initiation factor 4B; eIF4E, eukaryotic initiation factor 4E; eIF4G, eukaryotic initiation factor 4G; GSK3, glycogen synthase kinase 3; Mnk, MAPK-interacting kinase; mTORC1/2, mTOR complex 1/2; RSK 1/2, p90 ribosomal S6 kinase 1/2; PI3K, phosphatidylinositol 3-hydroxy kinase; pdcd4, programmed cell death 4; RTK, receptor tyrosine kinase; S6, ribosomal protein S6; S6K1, S6 kinase 1; TSC1/2, tuberous sclerosis 1/2.
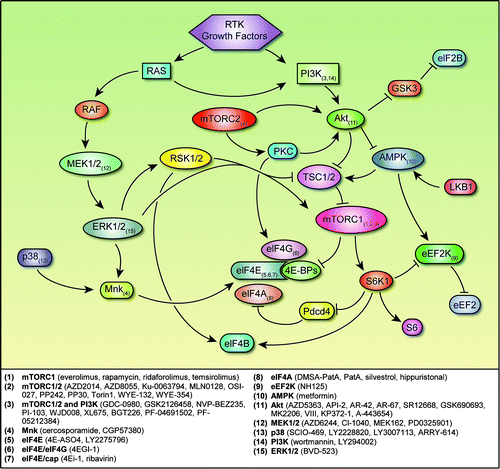
Translation elongation also is regulated by Akt/mTOR signaling.Citation13 In this process, a charged tRNA docks onto the ribosome/mRNA complex and adds amino acids into the growing polypeptide, and the process is repeated.Citation16 Further down, S6K1 phosphorylates and inhibits eukaryotic elongation factor 2 (eEF2) kinase (eEF2K), which is a negative regulator of eEF2, which catalyzes the translocation of the tRNA from the A-site to the P-site of the ribosome. S6 proposed to phosphorylate and regulate both the activity and the protein stability of programmed cell death 4 (pdcd4).Citation17-Citation19 In addition, S6K1 regulates estrogen receptor α activation.Citation20 Overall, S6K1 promotes tumor progression suggesting additional involvement of Akt/mTOR signaling in poor outcomes in cancer.
Akt/mTOR signaling also regulates ribosome biosynthesis,Citation13 which involves transcription and folding of pre-rRNAs, the folding and assembly or these RNAs with ribosomal proteins, and transport of these complexes to the cytoplasm for maturation. Akt signaling regulates ribosomal biogenesis by controlling the synthesis of rRNAs and ribosomal proteins. However, it is not clear whether activated Akt signaling increases ribosomal biogenesis and in turn increases protein synthesis.Citation13
Lastly, another mechanism of translation initiation, internal ribosome entry site–mediated translation, may also be regulated by Akt. This mechanism uses secondary RNA structures called internal ribosome entry sites (which are located mostly in the 5′ untranslated region of mRNA) to initiate translation in an eIF4E- and cap-independent manner and is thought to take part in translation of key mRNAs under conditions such as hypoxia, starvation, and DNA damage–inducing therapy.Citation21 These mRNAs encode proteins such as BCL-2, c-MYC, hypoxia-inducible factor–1α, vascular endothelial growth factor, and XIAP.Citation22-Citation26
Translational Control in Breast Cancer and Other Cancers
A significant amount of data suggests a relationship between translational control and breast cancer. A recent study showed that alterations in several translational regulators were associated with clinical-pathologic factors and with survival outcomes in hormone receptor–positive breast cancer.Citation27 High eEF2, S6, phosphorylated S6, and phosphorylated 4E-BP1 (p-4E-BP1) levels were associated with lymph node metastasis. High 4E-BP1 and p-4E-BP1 levels were associated with poor relapse-free survival. High p-4E-BP1, high phosphorylated S6, and low pdcd4 levels were associated with poor overall survival. In multivariate analysis, lymph node metastasis and high p-4E-BP1 level were predictors of shorter relapse-free survival, and high phosphorylated S6, high eEF2K, and low pdcd4 were associated with poor overall survival. Thus, this study confirmed an association between deregulation of translation and worse breast cancer outcomes. These results are consistent with those of another study, where PI3K pathway activation predicted poor outcomes after endocrine therapy in 64 patients with hormone receptor–positive breast cancer.Citation28
eIF3 subunits
In mammals, eIF3 is the largest scaffolding initiation factor, made up of 13 proteins.Citation3 It is involved in bridging the interaction between the 43S pre-initiation complex and eIF4F-bound mRNA. Several of its subunits take part in global protein expression, whereas others are involved in regulation of the eIF3 complex or interaction with specific mRNAs.
Alterations in expression of the eIF3 subunits -a, -b, -c, -h, and -i have been associated with oncogenic properties, such as increased cell growth, in vitro.Citation29 Of these, expression subunits -a, -c, and -h are increased in human cancers. For instance, eIF3a is overexpressed in breast, lung, cervix, stomach and esophagus cancers.Citation30,Citation31 eIF3a regulates mRNAs that inhibit the cell cycle and is required to maintain the malignant phenotype.Citation32,Citation33 eIF3c is overexpressed in seminomas and meningiomas.Citation34,Citation35 The oncogenic properties associated with eIF3c overexpression, such as high proliferation, are thought to result from the activation of translation and protein synthesis and the inactivation of the tumor suppressor merlin.Citation29,Citation35 In addition, in 18% of untreated primary breast tumors, EIF3H was amplified.Citation36 The chromosomal location of EIF3H is adjacent to that of the proto-oncogene MYC, and they are frequently amplified together.
Conversely, eIF3f expression is suppressed in several cancers, including breast cancer.Citation37 Overexpression of eIF3f inhibits cell proliferation, induces apoptosis, and inhibits translation and protein expression.Citation37 eIF3e binds both to eIF4G and to 40S, thus generating an interaction between the eIF4F complex and the ribosome.Citation38 Several studies support a tumor suppressor role for eIF3e. Increased expression of truncated eIF3e induces malignant transformation of normal mammary cells in vitro.Citation39 The EIF3E gene was found to have reduced expression in 37% and loss of heterozygosity in 21% of human breast cancers, implicating a loss of EIF3 activity in the development and progression of breast cancer.Citation40 In tamoxifen-treated breast cancer patients, high levels of EIF3E were significantly associated with prolonged progression-free survival and with therapy resistance.Citation41 A recent study reported that decreased expression of eIF3e induced epithelial-mesenchymal transition, invasive and migratory properties in breast epithelial cells, and an increase in the expression of key epithelial-mesenchymal transition regulators.Citation42 However, the loss of eIF3e expression reportedly did not significantly decrease global protein synthesis, although it did increase translation of mRNAs involved in cell proliferation, invasion, and apoptosis.Citation43,Citation44 Also, in primary breast cancer patients, eIF3e expression correlated positively with tumor grade.Citation44 These findings imply that eIF3e upregulates translation of oncoproteins, which conflicts with its tumor suppressor role. The exact function of eFI3e in breast cancer is not clear yet.
eIF4A
mTORC1 can activate cap-dependent translation by phosphorylating S6K. Activated S6K results in degradation of pdcd4, which is an eIF4A inhibitor, thus enhancing eIF4A activity.Citation17 The eIF4A helicase in turn initiates translation by unwinding highly structured 5′ untranslated regions in mRNAs.Citation45 eIF4A is overexpressed in primary hepatocellular carcinomasCitation46 and melanoma cell linesCitation47; beyond this, the role of eIF4A in cancer has not been well documented.
The overexpression of Mucin 1 (MUC1) in transgenic mice is associated with the formation of breast tumors.Citation48 MUC1 C-terminal (MUC1-C) subunit interacts with epidermal growth factor receptor (EGFR) and other receptor tyrosine kinases.Citation49 Translation MUC1-C is regulated by mTOR/S6K/programmed cell death protein 4 (pdcd4), which is an inhibitor of eIF4A.Citation50 Growth factor stimulation of MUC1 expression in vitro is inhibited by the eIF4A inhibitors silvestrol and CR-1–31-B, which points to eIF4A as a potential target in the treatment of breast cancer in patients with high MUC1 expression.Citation50 Furthermore, silvestrol has shown antitumor activity in mouse lymphoma and leukemia models, supporting the use of eIF4A inhibition for cancer treatment.Citation51-Citation53
eIF4E, eIF4G, and 4E-BPs
The most well-known oncogenic alteration in translation is overexpression of eIF4E. The availability of eIF4E is controlled by its binding with 4E-BPs and its phosphorylation by MAPK-interacting kinases (Mnks) 1 and 2.Citation54,Citation55 However, eIF4E expression and phosphorylation levels in nontransformed mammary epithelial cells and in breast cancer cells do not significantly differ; instead, eIF4GI protein expression and 4E-BP1 phosphorylation were greater in the transformed cells than in the nontransformed cells.Citation56 Thus, breast cancer cells can use not only eIF4E but also eIF4G and phosphorylation of 4E-BP1 to activate the eIF4F complex.
Increased activity of eIF4E regulates the selective expression of proteins such as cyclin D1, Bcl-2, Bcl-xL, and vascular endothelial growth factor by boosting nucleocytoplasmic transport for selected mRNAs, such as cyclin D1, and upregulating Nijmegen breakage syndrome protein 1 to activate Akt.Citation57-Citation63 These proteins are involved in cellular growth, angiogenesis, survival, and tumorigenesis. Until recently, this selective translation of mRNAs was not explained thoroughly. In addition to cap binding, eIF4E has been shown to stimulate eIF4A helicase by binding to eIF4GCitation64; the eIF4E/eIF4G binding interaction throughout scanning likely explains the selective translation of highly structured mRNAs.Citation64
The association between eIF4E and cancer has been demonstrated preclinically and clinically observed. eIF4E overexpression has contributes to the transformation of fibroblasts and mammary epithelial cells, enabling uncontrolled cell proliferation.Citation65 In transgenic mice, eIF4E overexpression initiates the formation of tumors with various histologies, including B-cell lymphomas, angiosarcomas, lung adenocarcinomas, and hepatocellular adenomas.Citation66 Also, ectopic expression of eIF4E in vitro repairs the defect in eIF4F formation due to drugs targeting HER2 and EGFR, whereas high eIF4E expression s been associated with incomplete response in patients receiving anti-HER therapies, and thus is a potential marker of resistance.Citation67 eIF4E overexpression has been shown in breast, bladder, cervix, colorectal, head and neck, lung, prostate, and skin cancers, as well as lymphomas.Citation3,Citation57 In breast cancer, high eIF4E has been suggested as an indicator of increased cancer recurrence and risk of death, and several clinical studies show that high eIF4E indicates a poor prognosis independently of nodal or HER2 status.Citation15,Citation68-Citation72
Another mediator of translation initiation is the binding and unbinding of eIF4E by 4E-BPs, which compete with eIF4G for a eIF4E binding site. Hyperphosphorylation of 4E-BPs decreases this binding and inactivates the 4E-BPs, increasing eIF4E availability to engage in the cap-complex and to activate eIF4F. Conversely, phosphorylated 4E-BPs bind to eIF4E and remove eIF4G, thereby inhibiting translation initiation.
Low total 4E-BP1 levels in prostate cancer patients were associated with shorter survival than normal 4E-BP1 expression was.Citation73 Similarly, higher p-4E-BP1 levels in advanced prostate cancer were associated with shorter survival.Citation73 In breast cancer, increased eIF4E and p-4E-BP1 levels correlate with higher tumor grade and shorter survival, whereas increased expression of total 4E-BP1 and 4E-BP2 is associated with lower tumor grade.Citation74 In these studies, both the loss of 4E-BP1 expression, and increased p-4E-BP1 and eIF4E levels indicate poor prognosis.
However, contradicting data show that total 4E-BP1 and eIF4G are overexpressed in large advanced breast cancers.Citation75 Under hypoxia, elevated levels of these two proteins inhibit cap-dependent translation and facilitate selective translation of mRNAs that contain internal ribosome entry sites, promoting tumor angiogenesis and growth.Citation75
In a recent study, protein phosphatase PPM1G dephosphorylated 4E-BP1 and negatively regulated protein translation in vitro.Citation76 A time course experiment showed that 4E-BP1 dephosphorylation by amino acid starvation or by mTOR inhibition was significantly delayed in PPM1G knockdown cells.Citation76 It is not yet known whether PPM1G expression affects the prognosis of cancer patients.
Like eIF4E overexpression, overexpression of eIF4G1 in vitro leads to malignant transformation.Citation77 eIF4G may contribute to this transformation by promoting cap-dependent translation by forming active eIF4F or by increasing translation of internal ribosome entry site–dependent mRNAs.Citation4 In contrast, decreased eIF4G1 expression impairs cell proliferation and bioenergetics and promotes autophagy by inhibiting translation of mRNAs.Citation78 The significant overexpression of eIF4G and 4E-BP1 in locally advanced breast cancers was shown to result in a hypoxia-activated shift from cap-dependent translation to cap-independent translation.Citation75 This shift results in cap-independent translation of mRNAs such as vascular endothelial growth factor (pro-angiogenic), hypoxia-inducible factor–1α (hypoxia stress–induced), and Bcl2 (survival-promoting), increasing tumor angiogenesis and local tumor growth. Indeed, in inflammatory breast cancer, most tumors show overexpression of eIF4G1 without any change in levels of eIF4E and 4E-BP1.Citation79 eIF4G1 also promotes cap-independent translation of mRNAs that are responsible for tumor cell emboli and invasion.
The eIF4F complex and its components eIF4E, eIF4G, and eIF4A are widely studied and are a major focus of therapeutic interventions.Citation5,Citation63 New inhibitors that target various mechanisms within this complex are emerging. These developments should provide new insights into translational control in cancer and should open new therapeutic possibilities.
Translation Elongation via eEF2K and eEF2
Many signaling pathways control eEF2K activity, and these signals are relayed through eEF2. For example, MAPK and mTOR pathways inhibit eEF2K in response to nutrient and growth factor signals, whereas adenosine monophosphate–activated protein kinase, PKA, and calmodulin activate eEF2K in response to energy- and Ca2+-dependent signaling.Citation80 eEF2K downregulates eEF2 to inhibit protein synthesis when conservation of resources is necessary. eEF2K/eEF2 signaling may promote cancer cell survival under stress conditions, such as starvation, hypoxia, or autophagy.Citation81 The activity of eEF2 is increased in several cancer cell lines and tumor types including ovarian, gastric, and colon cancer.Citation82-Citation84 In gastric cancer cell lines, eEF2 overexpression promotes progression in the cell cycle, inactivates eEF2K, and activates Akt and cdc2.Citation84 eEF2K upregulates autophagy, and its inhibition activates apoptosis.Citation81,Citation85 eEF2K is upregulated in malignant rat glial cells and in human melanoma and breast cancer.Citation86-Citation89 In a mouse model of triple-negative breast cancer, systemic administration of eEF2K siRNA downregulated eEF2K expression and eEF2 phosphorylation, leading to inhibition of tumor growth.Citation90 Overexpression of these proteins in cancer makes them targets for treatment.
Additional Translation Mechanisms
Pdcd4
Pdcd4 is a tumor suppressor protein that inhibits cancer cell invasion, and its expression is often decreased in breast cancer.Citation91 It inhibits translation of specific mRNAs, with downstream effects on cell signaling and oncogenic mRNA transcription.Citation92 Pdcd4 also binds eIF4A and thus inhibits the formation and function of the eIF4F complex.Citation93 It is downregulated by tumor promoters and undergoes degradation by β-TrCP after phosphorylation by S6K1.Citation17,Citation94
Unfolded protein response
Endoplasmic reticulum stress induces selective translation in cancer cells. Unfolded protein response heals the damage from this stress and restores endoplasmic reticulum homeostasis. To reduce the influx of nascent peptides into overloaded endoplasmic reticulum, a protein kinase, PERK, phosphorylates eIF2.Citation95 This phosphorylation also aids translation of stress-related mRNAs.Citation95 Glucose-related protein 78 has a central role in the unfolded protein response; it is overexpressed in many cancers and usually associated with poor response.Citation96
MicroRNAs
MicroRNAs are non-coding RNAs that play important roles in all biologic pathways. They also regulate several cancer-related processes such as proliferation and apoptosis.Citation97 Several studies address the relationships between microRNAs and cancer and between microRNAs and translation initiation.Citation97,Citation98
The roles of the unfolded protein response and microRNAs in cancer are beyond the scope of this review, but considering the important steps that they control, they are potential platforms for development of novel cancer treatments.
Targeting of Proteins Involved in Protein Synthesis
Several approaches to altering the rate of translation are in development, pursuing translation initiation as a therapeutic target. Of the factors that are involved in translational control and summarized above, eIF4E and 4E-BPs are prominent potential targets. eIF4E is required for cap-dependent translation; however, decreased eIF4E levels appear to have no effect on normal tissues, rendering cancer tissue more susceptible to targeting of this factor.Citation99 siRNA knockdown of eIF4E inhibits cancer cell growth in a variety of breast cancer cell lines.Citation100 In addition, treatment with second-generation antisense oligonucleotides targeting eIF4E (4E-ASO4) reduces tumor growth in nude mice bearing human prostate and breast cancer xenografts.Citation101 A phase I trial of the eIF4E-targeting antisense oligonucleotide LY2275796 was recently completedCitation102; the treatment was well tolerated and tumor eIF4E expression suppressed, but no objective tumor responses were observed. The efficacy of eIF4E antisense oligonucleotides in combination with chemotherapy is still being evaluated in clinical trials.Citation103,Citation104
eIF4E inhibitor 1, a cap analog competing with the cap structure for binding to eIF4E, inhibited cap-dependent translation in zebrafish embryos and prevented eIF4E-initiated epithelial-mesenchymal transition in zebrafish ectoderms.Citation105 Another physical mimic of the 7-methylguanosine cap structure, ribavirin, competes with eIF4E binding and disrupts transport and translation of mRNAs regulated by eIF4E.Citation99 Of 11 acute myeloid leukemia cases treated with ribavirin, there was one complete remission and two partial remissions, and reduced eIF4E levels after the treatment were associated with clinical response.Citation106 Unfortunately, these responses were transient, and all patients developed resistance in months.Citation107 Most of the following trials of ribavirin in acute myeloid leukemia combined ribavirin with cytotoxic chemotherapy, such as cytosine arabinoside.Citation107-Citation109 Ribavirin also was given to metastatic breast cancer patients as a single agent in a recently terminated phase I/II study.Citation110 Despite the presented evidence on the use of ribavirin in treating human cancer, some studies question its role as a functional mimic of the mRNA cap in vitro.Citation111,Citation112
Peptides based on the eIF4E-binding motifs of 4E-BPs and eIF4G interfere with eIF4E-eIF4G binding, translation initiation, the cell cycle, and survival in vitro.Citation113,Citation114 A 4E-BP–based peptide fused to an analog of gonadotropin-releasing hormone is taken up by ovarian tumor cells expressing the gonadotropin-releasing hormone receptor and inhibits growth in vitro and in vivo.Citation115 A synthetic peptide, 4EGI-1, binds eIF4E and inhibits eIF4E/eIF4G interaction,Citation116 thus inhibiting cap-dependent translation, increasing eIF4E–4E-BP1 association, and decreasing expression of oncogenic proteins. The use of another synthetic peptide, 4E1RCat, to interfere with the eIF4E-eIF4G interaction was reported to reverse tumor resistance in a lymphoma mouse model.Citation117
Pdcd4 inhibits protein translation by binding to eIF4A.Citation93 Pdcd4 also inhibits Jun kinase activation, AP-1–dependent transcription, transformation, tumorigenesis, and invasion.Citation94 In at least some cancer cell lines, PI3K/mTOR inhibitors can be used to increase pdcd4 expression. However, an alternative strategy is to target eIF4A directly. The natural product pateamine A is a small-molecule modulator of eIF4A activity and an inhibitor of translation.Citation118 Pateamine A causes eIF4A to be sequestered on RNA and limits eIF4A availability for incorporation into the eIF4F complex.Citation119 Des-methyl, des-amino pateamine A is a structurally simplified analog of pateamine A and shows potent antiproliferative activity in a variety of human cancer cell lines and xenograft models.Citation120 Silvestrol is a compound with a similar mechanism, promoting interaction between eIF4A and capped RNA, interfering with the assembly of eIF4F complex, and thus inhibiting translation initiation.Citation51,Citation121 This drug was shown to inhibit translation and anticancer activity in breast and prostate cancer xenograft models and in B-cell leukemias in vitro and in vivo.Citation52,Citation121 Hippuristanol, a natural eIF4A RNA-binding activity inhibitor, induced cell cycle arrest and apoptosis in vitro and reduced tumor growth in vivo.Citation122,Citation123
N1-guanyl-1,7-diaminoheptane and ciclopirox olamine are inhibitors of deoxyhypusine synthase and deoxyhypusine hydroxylase, respectively.Citation124,Citation125 These agents inhibit hypusination of eIF5A and reduce proliferation in various cancer cell lines and inhibit tumor growth in xenograft models of human breast cancer, melanoma, and leukemia.Citation126-Citation128
eEF2K integrates a variety of signals and thus may be a target for different strategies. For example, insulin and serum stimulation activates eEF2K and inhibits eEF2; these effects are blocked by rapamycin.Citation129 Another study showed that high doses of the rapamycin analog temsirolimus reduced S6K1 and eEF2K phosphorylation but, unexpectedly, increased eEF2 phosphorylation on Thr56, the eEF2K phosphorylation site.Citation130 This increase involved an S6K1-independent mechanism, as it did not correlate with decreased S6K1 activity or with decreased eEF2K phosphorylation. Additional studies are needed to determine the effect of clinically achievable doses of PI3K/mTOR inhibitors in vivo and their effect on hormone receptor–positive breast cancers that express low or high eEF2K. NH125, a 2-methyl-imidazolium derivative, is a potent and relatively specific eEF2K inhibitorCitation82; a panel of cell lines, including a breast cancer cell line, that were treated with NH125 showed inhibited cell growth.
Protein elongation is also targeted by omacetaxine mepesuccinate (formerly homoharringtonine). This agent prevents aminoacyl-tRNA binding to the ribosomal acceptor site and peptide bond formation.Citation131,Citation132 It competes with amino acid side chains of incoming aminoacyl-tRNAs for binding at the A-site cleft in the peptidyl transferase center.Citation133 Omacetaxine induces loss of proliferation and survival proteins, such as Mcl-1, cyclin D1, c-Myc, in a cell lines of chronic myeloid leukemia, acute myeloid leukemia, chronic lymphocytic leukemia and multiple myeloma. Only cells that rely on expression of these proteins are likely to be affected by omacetaxine, in other cancer types it is not sufficient to induce apoptosis.Citation134
Targeting of Signaling Networks Involved in Protein Synthesis in Clinical Trials
The PI3K/Akt/mTOR pathway has essential roles in normal cellular function. Akt and its downstream effectors control cell growth and survival, and mTOR participates in biologic processes and regulates protein synthesis. The activation of mTOR is driven by mitogens through PI3K/Akt. Alternatively, mTOR can be activated through the Ras/MEK/ERK pathway.Citation135 The prototypical allosteric mTOR inhibitor is rapamycin. Its analogs (also called rapalogs) are everolimus, ridaforolimus, and temsirolimus. These agents primarily inhibit mTORC1. In addition to translation inhibition, they induce cell cycle arrest and sensitize cancer cells to chemotherapeutic agents.Citation136 Since the PI3K/Akt/mTOR pathway is frequently activated in cancer, several clinical trials are testing the therapeutic effect of combining rapamycin analogs with other inhibitors.
In a phase III trial, 626 patients with previously untreated, poor-prognosis metastatic renal carcinoma were randomized into temsirolimus, interferon α or combination therapy groups.Citation137 The temsirolimus group had longer overall survival (hazard ratio [HR], 0.73; 95% confidence interval [95% CI], 0.58–0.92; P = 0.008) and progression-free survival (PFS) (P < 0.001) than the interferon group. The median overall survival times for the interferon, temsirolimus, and combination groups were 7.3, 10.9, and 8.4 mo, respectively. Another phase III trial compared everolimus and placebo treatment in patients with previously treated progressive metastatic renal cell carcinoma.Citation138 The trial was stopped early because of the significant efficacy of everolimus (HR, 0.30; 95% CI, 0.22–0.40; P < 0.0001). The median PFS times were 4.0 mo in the everolimus group and 1.9 mo in the placebo group. The RADIANT-3 trial randomized 410 patients with advanced pancreatic neuroendocrine tumors to receive everolimus or placebo, and both with best supportive care.Citation139 The median PFS times were 11.0 mo with everolimus and 4.6 mo with the placebo (HR, 0.35; 95% CI, 0.27–0.45; P < 0.001). Everolimus decreased the estimated risk of progression or death by 65%, thus significantly prolonging PFS. In the EXIST-1 trial, 117 patients with subependymal giant cell astrocytomas associated with tuberous sclerosis received either everolimus or a placebo.Citation140 At least a 50% reduction in tumor volume was observed in 35% of the patients in the everolimus group and 0% of the patients on the placebo (P < 0.0001). In the EXIST-2 study, 118 patients with angiomyolipoma and tuberous sclerosis or lymphangioleiomyomatosis were assigned into everolimus or placebo groups.Citation141 At least a 50% reduction in tumor volume was observed in 42% of the patients in the everolimus group and 0% of patients in the placebo group (P < 0.0001).
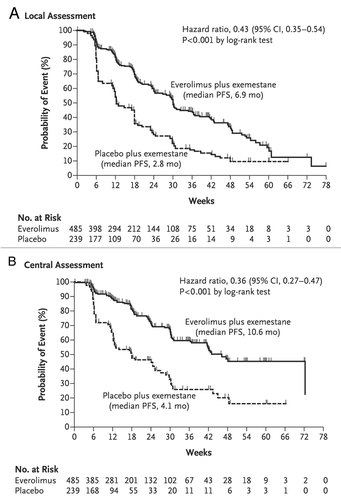
Several clinical trials have been conducted using mTOR inhibitors in breast cancer. Of 33 patients with metastatic breast cancer who received daily everolimus as a single agent, 12% showed a response (95% CI, 3.4–28.2%).Citation142 Most estrogen receptor–positive patients benefit from aromatase inhibitors; however, most of them eventually develop progression or recurrence. Estrogen-depleted cells rely on activation of mTOR signalingCitation28; as a result, inhibition of mTOR signaling sensitizes these cells to aromatase inhibitors.Citation143 Thus, there is cross-talk between estrogen receptor signaling and PI3K/Akt/mTOR signaling, and the latter is responsible for the development of resistance to aromatase inhibitors. Striking results were seen in a phase II study of the combination of the estrogen receptor antagonist tamoxifen with everolimus in comparison with tamoxifen alone in patients with hormone receptor–positive, epidermal growth factor receptor 2–negative, aromatase inhibitor–naïve metastatic breast cancer.Citation144 The 6-mo rate of clinical benefit was 61% with the combination and 42% with tamoxifen alone, and the risk of death was 55% lower with combination therapy than with tamoxifen alone. In another phase II trial, 270 postmenopausal women with operable estrogen receptor–positive breast cancer received neoadjuvant treatment with letrozole (a nonsteroidal aromatase inhibitor) with or without everolimus. Greater inhibition of cell proliferation and better clinical response rates were observed in the combination arm (response rates of 68.1% with letrozole alone and 59.1% with the combination; P = 0.062, one-sided α = 0.1).Citation145 The aromatase inhibitor exemestane inhibits estrogen synthesis. Recently, the BOLERO-2 phase III trial included 724 patients with hormone receptor–positive advanced breast cancer that had previously been treated with nonsteroidal aromatase inhibitors and had progression or recurrence, and/or with advanced disease.Citation146,Citation147 Patients were assigned into exemestane and exemestane plus everolimus groups. According to an assessment by local investigators, median PFS times were 6.9 mo in the combination group and 2.8 mo in the exemestane-only group (HR, 0.43; 95% CI, 0.35–0.54; P < 0.001) (). According to an assessment by an independent committee, the median PFS times were 10.6 mo for the combination and 4.1 mo for exemestane only (HR, 0.36; 95% CI, 0.27–0.47; P < 0.001) (). Thus, adding everolimus to exemestane improved PFS in these patients. A phase III study that was similarly planned but targeted nonsteroidal aromatase inhibitor–naïve patients with temsirolimus was terminated owing to a lack of responses at the interim analysis.Citation148 Letrozole alone or with temsirolimus was offered as a first-line therapy to 1,112 patients; no difference in PFS between the groups was observed.
Figure 2. Kaplan-Meier estimates of progression-free survival rates. (A) Using radiographic studies, local investigators assessed the primary end point, progression-free survival. (B) A second assessment was conducted by an independent radiology committee. Reprinted from the New England Journal of Medicine 2012;366:520–9, with permission.
ATP-competitive inhibitors of mTOR—the mTOR kinase inhibitors—are potent inhibitors of mTORC1 and mTORC2; these inhibitors include Ku-0063794, PP30, Torin1, WYE-132, and WYE-354 in preclinical settings and AZD2014, AZD8055, MLN0128, OSI-027, and PP242 in clinical trials. They more effectively inhibit mTOR preclinical studies, resulting in dephosphorylation of 4E-BPs and impairment of translation initiation. They also inhibit Akt phosphorylation by directly inhibiting mTORC2. Clinical trials are ongoing to determine the efficacy of mTOR kinase inhibitors as single agents and in combination therapy. Lastly, there are PI3K/mTOR dual inhibitors, which are more effective than rapamycin but also may be more toxic. PI-103 and WJD008have been used in preclinical studies.Citation149,Citation150 XL765 GDC-0980, GSK2126458, NVP-BEZ235, PF-04691502, PF-05212384, and LY3023414 are being tested in clinical trials. These inhibitors are more efficient than mTOR inhibitors in blocking Akt phosphorylation.Citation151
Akt is associated with tumor cell survival, proliferation, and invasiveness, and its activation is one of the most frequent alterations in cancer. Akt directly phosphorylates and inactivates tuberous sclerosis 2, and tuberous sclerosis 2 was shown to inhibit S6K1 and activate 4E-BP1 through mTOR.Citation152 Thus, activated Akt stimulates mTOR and promotes activation of its downstream targets. Akt inhibitors API-2, AR-42, AR-67, and SRI2668 have been tested only in healthy subjects, and AZD5363, and GSK690693 are being tested in clinical trials. Also, the Akt inhibitor MK2206 is being evaluated in patients with advanced breast cancer and in the neoadjuvant setting in patients with operable breast cancer.Citation153,Citation154 Other Akt-inhibiting compounds, such as VIII, KP372–1, and A-443654, are being used in preclinical studies.
Approximately 25% of breast cancers overexpress HER2, which is associated with poor prognosis in patients not treated with HER2-targeted therapy.Citation155 HER2 activates both the PI3K/Akt/mTOR and the RAS/MAPK pathways—one of several connections between those pathways. Rapamycin inhibition of mTORC1 was shown to activate an S6K-PI3K-Ras feedback loop, leading to MAPK activation.Citation156 In addition, oncogenic MAPK signaling activates mTORC1 through the RSK1/2-raptor and ERK1/2-raptor axes.Citation157,Citation158 Inhibitors of the RAS/MAPK pathway component MEK1/2, including AZD6244, Cl−1040, MEK162, PD0325901, and MEK162, are very promising in the treatment of solid cancer. MEK162 has been evaluated in phase III trials, and trametinib has been approved by the FDA for Raf-mutant metastatic melanoma. These two agents are being tested for the treatment of breast cancer in phase I trials: trametinib as a single agent or in combination therapy and MEK162 in combination therapy.
The regulation of eIF4E forms another node of convergence between the PI3K/Akt/mTOR and RAS/MAPK signaling pathways. First, the PI3K/Akt/mTOR pathway phosphorylates 4E-BPs, liberates eIF4E, and allows eIF4F formation. Second, MAPK-interacting kinases 1 and 2 phosphorylate eIF4E, a modification that is required for eIF4E-dependent cell survival. Thus, mTOR and Mnk inhibition may inhibit cell proliferation. Indeed, in a panel of six cell lines, five showed a reduction in proliferation in response to treatment with Mnk kinase inhibitor N3-(4-fluorophenyl)-1H-pyrazolo-[3,4-d]pyrimidine-3,4-diamine (CGP57380).Citation159 The only unresponsive cell line had no detectable baseline eIF4E phosphorylation. The antifungal agent cercosporamide is an orally bioavailable Mnk inhibitor, particularly of Mnk2.Citation160 In cancer cell lines of various origins, cercosporamide blocked proliferation; mouse experiments showed dephosphorylation of eIF4E 30 min after cercosporamide administration. Thus, the published literature suggests that suppressing Mnk function has potential in cancer treatment, like other approaches inhibiting eIF4E activity.
Obesity and diabetes are associated with increased breast cancer risk.Citation161 Metformin is an anti-diabetic drug that exerts anti-cancer effects. For instance, patients with breast cancer and diabetes on metformin and neoadjuvant chemotherapy have a higher pathologic complete response rate than either breast cancer patients with diabetes receiving neoadjuvant chemotherapy but not metformin or nondiabetic breast cancer patients.Citation162 The insulin-independent mode of action is not clear, but metformin activates adenosine monophosphate–activated protein kinase, which inhibits the mTOR pathway.Citation163 The dose of metformin needed to inhibit mTOR is not known; however, in vitro co-administration of metformin and everolimus with chemotherapeutic agents increased inhibition of cell proliferation.Citation164 A meta-analysis seven clinical studies assessed whether metformin is associated with decreased breast cancer risk and suggested that metformin is protective among postmenopausal breast cancer patients with diabetes, these patients have a lower risk of developing invasive breast cancer.Citation165 A neoadjuvant window of opportunity study of metformin in women with newly diagnosed, untreated, non-diabetic breast cancer who were waiting for surgery resulted in significant decreases in insulin, leptin, and C-reactive protein levels, a decrease in Ki67 staining of tumor tissue, and an increase in terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling staining.Citation166 These cellular changes were consistent with the anti-cancer effect of metformin. Several clinical trials are evaluating metformin administration in combination treatments for early and metastatic breast cancer. Further research is needed to identify the mechanisms of metformin’s anti-tumor action and which cancer patients will benefit from metformin treatment.
Omacetaxine mepesuccinate was recently approved by FDA to treat chronic myeloid leukemia patients for resistance or tolerance to two or more tyrosine kinase inhibitors.Citation167,Citation168 Data pooled from two phase 2 trials showed hematologic response in 69% and major cytogenetic response in 20% of patients.Citation168 Median duration of hematologic response and major cytogenetic response were 12.2 mo and 17.7 mo, respectively. This is a novel approach to treat patients with chronic myeloid leukemia, and clinical trials are evaluating omacetaxine for different types of cancer and in drug combinations.
Conclusion
Emerging data show the importance of the dysregulation of translation in the development and progression of cancer. The first step of translation, translation initiation, appears to have a significant role in many cancer lineages, including breast cancer. Several agents target components of translation initiation, and many other agents target the components of the signaling pathways that regulate translation initiation. Most of these agents are in clinical trials, and some already have been approved for clinical use in cancer. As single agents or in combination treatments, they can inhibit tumor growth and prevent or overcome resistance to treatment. Further clinical studies will allow us to determine the best approach to personalizing treatment in breast cancer patients with translational dysregulations.
Disclosure of Potential Conflicts of Interest
The authors have no disclosures.
Acknowledgments
Dr. Meric-Bernstam’s laboratory was supported by a Society of Surgical Oncology Clinical Investigator Award in Breast Cancer Research sponsored by Susan G. Komen for the Cure, Susan G. Komen for the Cure grant SAC10006, Stand Up to Cancer Dream Team Translational Research Grant, a Program of the Entertainment Industry Foundation grant, a Kleberg Foundation grant, National Center for Research Resources grants 3UL1RR024148 and UL1TR000371, and the National Cancer Institute through R21CA159270 and The University of Texas MD Anderson Cancer Center Support Grant (P30CA016672).
References
- American Cancer Society. Cancer Facts and Figures 2013. Atlanta, GA: American Cancer Society, 2013.
- Meric F, Hunt KK. Translation initiation in cancer: a novel target for therapy. Mol Cancer Ther 2002; 1:971 - 9; PMID: 12481419
- Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nat Rev Cancer 2010; 10:254 - 66; http://dx.doi.org/10.1038/nrc2824; PMID: 20332778
- Grzmil M, Hemmings BA. Translation regulation as a therapeutic target in cancer. Cancer Res 2012; 72:3891 - 900; http://dx.doi.org/10.1158/0008-5472.CAN-12-0026; PMID: 22850420
- Malina A, Cencic R, Pelletier J. Targeting translation dependence in cancer. Oncotarget 2011; 2:76 - 88; PMID: 21378410
- Ruggero D. Translational control in cancer etiology. Cold Spring Harb Perspect Biol 2013; 5:5; http://dx.doi.org/10.1101/cshperspect.a012336; PMID: 22767671
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 2009; 136:731 - 45; http://dx.doi.org/10.1016/j.cell.2009.01.042; PMID: 19239892
- Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem 1999; 68:913 - 63; http://dx.doi.org/10.1146/annurev.biochem.68.1.913; PMID: 10872469
- Blagden SP, Willis AE. The biological and therapeutic relevance of mRNA translation in cancer. Nat Rev Clin Oncol 2011; 8:280 - 91; http://dx.doi.org/10.1038/nrclinonc.2011.16; PMID: 21364523
- Aitken CE, Lorsch JR. A mechanistic overview of translation initiation in eukaryotes. Nat Struct Mol Biol 2012; 19:568 - 76; http://dx.doi.org/10.1038/nsmb.2303; PMID: 22664984
- Wells SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell 1998; 2:135 - 40; http://dx.doi.org/10.1016/S1097-2765(00)80122-7; PMID: 9702200
- Meric-Bernstam F, Gonzalez-Angulo AM. Targeting the mTOR signaling network for cancer therapy. J Clin Oncol 2009; 27:2278 - 87; http://dx.doi.org/10.1200/JCO.2008.20.0766; PMID: 19332717
- Hsieh AC, Truitt ML, Ruggero D. Oncogenic AKTivation of translation as a therapeutic target. Br J Cancer 2011; 105:329 - 36; http://dx.doi.org/10.1038/bjc.2011.241; PMID: 21772331
- Meric-Bernstam F. Translation initiation factor 4E (eIF4E): prognostic marker and potential therapeutic target. Ann Surg Oncol 2008; 15:2996 - 7; http://dx.doi.org/10.1245/s10434-008-0116-7; PMID: 18712446
- Li BD, McDonald JC, Nassar R, De Benedetti A. Clinical outcome in stage I to III breast carcinoma and eIF4E overexpression. Ann Surg 1998; 227:756-6l; discussion 61-3.
- Dever TE, Green R. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb Perspect Biol 2012; 4:a013706; http://dx.doi.org/10.1101/cshperspect.a013706; PMID: 22751155
- Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science 2006; 314:467 - 71; http://dx.doi.org/10.1126/science.1130276; PMID: 17053147
- Wang X, Li W, Williams M, Terada N, Alessi DR, Proud CG. Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J 2001; 20:4370 - 9; http://dx.doi.org/10.1093/emboj/20.16.4370; PMID: 11500364
- Ferrari S, Bandi HR, Hofsteenge J, Bussian BM, Thomas G. Mitogen-activated 70K S6 kinase. Identification of in vitro 40 S ribosomal S6 phosphorylation sites. J Biol Chem 1991; 266:22770 - 5; PMID: 1939282
- Yamnik RL, Holz MK. mTOR/S6K1 and MAPK/RSK signaling pathways coordinately regulate estrogen receptor alpha serine 167 phosphorylation. FEBS Lett 2010; 584:124 - 8; http://dx.doi.org/10.1016/j.febslet.2009.11.041; PMID: 19925796
- Holcik M, Sonenberg N, Korneluk RG. Internal ribosome initiation of translation and the control of cell death. Trends Genet 2000; 16:469 - 73; http://dx.doi.org/10.1016/S0168-9525(00)02106-5; PMID: 11050335
- Sherrill KW, Byrd MP, Van Eden ME, Lloyd RE. BCL-2 translation is mediated via internal ribosome entry during cell stress. J Biol Chem 2004; 279:29066 - 74; http://dx.doi.org/10.1074/jbc.M402727200; PMID: 15123638
- Notari M, Neviani P, Santhanam R, Blaser BW, Chang JS, Galietta A, Willis AE, Roy DC, Caligiuri MA, Marcucci G, et al. A MAPK/HNRPK pathway controls BCR/ABL oncogenic potential by regulating MYC mRNA translation. Blood 2006; 107:2507 - 16; http://dx.doi.org/10.1182/blood-2005-09-3732; PMID: 16293596
- Lang KJ, Kappel A, Goodall GJ. Hypoxia-inducible factor-1alpha mRNA contains an internal ribosome entry site that allows efficient translation during normoxia and hypoxia. Mol Biol Cell 2002; 13:1792 - 801; http://dx.doi.org/10.1091/mbc.02-02-0017; PMID: 12006670
- Miller DL, Dibbens JA, Damert A, Risau W, Vadas MA, Goodall GJ. The vascular endothelial growth factor mRNA contains an internal ribosome entry site. FEBS Lett 1998; 434:417 - 20; http://dx.doi.org/10.1016/S0014-5793(98)01025-4; PMID: 9742966
- Holcik M, Yeh C, Korneluk RG, Chow T. Translational upregulation of X-linked inhibitor of apoptosis (XIAP) increases resistance to radiation induced cell death. Oncogene 2000; 19:4174 - 7; http://dx.doi.org/10.1038/sj.onc.1203765; PMID: 10962579
- Meric-Bernstam F, Chen H, Akcakanat A, Do KA, Lluch A, Hennessy BT, Hortobagyi GN, Mills GB, Gonzalez-Angulo AM. Aberrations in translational regulation are associated with poor prognosis in hormone receptor-positive breast cancer. Breast Cancer Res 2012; 14:R138; http://dx.doi.org/10.1186/bcr3343; PMID: 23102376
- Miller TW, Hennessy BT, González-Angulo AM, Fox EM, Mills GB, Chen H, Higham C, García-Echeverría C, Shyr Y, Arteaga CL. Hyperactivation of phosphatidylinositol-3 kinase promotes escape from hormone dependence in estrogen receptor-positive human breast cancer. J Clin Invest 2010; 120:2406 - 13; http://dx.doi.org/10.1172/JCI41680; PMID: 20530877
- Zhang L, Pan X, Hershey JW. Individual overexpression of five subunits of human translation initiation factor eIF3 promotes malignant transformation of immortal fibroblast cells. J Biol Chem 2007; 282:5790 - 800; http://dx.doi.org/10.1074/jbc.M606284200; PMID: 17170115
- Bachmann F, Bänziger R, Burger MM. Cloning of a novel protein overexpressed in human mammary carcinoma. Cancer Res 1997; 57:988 - 94; PMID: 9041205
- Yin JY, Shen J, Dong ZZ, Huang Q, Zhong MZ, Feng DY, Zhou HH, Zhang JT, Liu ZQ. Effect of eIF3a on response of lung cancer patients to platinum-based chemotherapy by regulating DNA repair. Clin Cancer Res 2011; 17:4600 - 9; http://dx.doi.org/10.1158/1078-0432.CCR-10-2591; PMID: 21610145
- Dong Z, Zhang JT. EIF3 p170, a mediator of mimosine effect on protein synthesis and cell cycle progression. Mol Biol Cell 2003; 14:3942 - 51; http://dx.doi.org/10.1091/mbc.E02-12-0784; PMID: 12972576
- Dong Z, Liu LH, Han B, Pincheira R, Zhang JT. Role of eIF3 p170 in controlling synthesis of ribonucleotide reductase M2 and cell growth. Oncogene 2004; 23:3790 - 801; http://dx.doi.org/10.1038/sj.onc.1207465; PMID: 15094776
- Rothe M, Ko Y, Albers P, Wernert N. Eukaryotic initiation factor 3 p110 mRNA is overexpressed in testicular seminomas. Am J Pathol 2000; 157:1597 - 604; http://dx.doi.org/10.1016/S0002-9440(10)64797-9; PMID: 11073819
- Scoles DR, Yong WH, Qin Y, Wawrowsky K, Pulst SM. Schwannomin inhibits tumorigenesis through direct interaction with the eukaryotic initiation factor subunit c (eIF3c). Hum Mol Genet 2006; 15:1059 - 70; http://dx.doi.org/10.1093/hmg/ddl021; PMID: 16497727
- Nupponen NN, Porkka K, Kakkola L, Tanner M, Persson K, Borg A, Isola J, Visakorpi T. Amplification and overexpression of p40 subunit of eukaryotic translation initiation factor 3 in breast and prostate cancer. Am J Pathol 1999; 154:1777 - 83; http://dx.doi.org/10.1016/S0002-9440(10)65433-8; PMID: 10362802
- Shi J, Kahle A, Hershey JW, Honchak BM, Warneke JA, Leong SP, Nelson MA. Decreased expression of eukaryotic initiation factor 3f deregulates translation and apoptosis in tumor cells. Oncogene 2006; 25:4923 - 36; http://dx.doi.org/10.1038/sj.onc.1209495; PMID: 16532022
- LeFebvre AK, Korneeva NL, Trutschl M, Cvek U, Duzan RD, Bradley CA, Hershey JW, Rhoads RE. Translation initiation factor eIF4G-1 binds to eIF3 through the eIF3e subunit. J Biol Chem 2006; 281:22917 - 32; http://dx.doi.org/10.1074/jbc.M605418200; PMID: 16766523
- Rasmussen SB, Kordon E, Callahan R, Smith GH. Evidence for the transforming activity of a truncated Int6 gene, in vitro. Oncogene 2001; 20:5291 - 301; http://dx.doi.org/10.1038/sj.onc.1204624; PMID: 11536042
- Marchetti A, Buttitta F, Pellegrini S, Bertacca G, Callahan R. Reduced expression of INT-6/eIF3-p48 in human tumors. Int J Oncol 2001; 18:175 - 9; PMID: 11115556
- Umar A, Kang H, Timmermans AM, Look MP, Meijer-van Gelder ME, den Bakker MA, Jaitly N, Martens JW, Luider TM, Foekens JA, et al. Identification of a putative protein profile associated with tamoxifen therapy resistance in breast cancer. Mol Cell Proteomics 2009; 8:1278 - 94; http://dx.doi.org/10.1074/mcp.M800493-MCP200; PMID: 19329653
- Gillis LD, Lewis SM. Decreased eIF3e/Int6 expression causes epithelial-to-mesenchymal transition in breast epithelial cells. Oncogene 2013; 32:3598 - 605; http://dx.doi.org/10.1038/onc.2012.371; PMID: 22907435
- Bandyopadhyay A, Matsumoto T, Maitra U. Fission yeast Int6 is not essential for global translation initiation, but deletion of int6(+) causes hypersensitivity to caffeine and affects spore formation. Mol Biol Cell 2000; 11:4005 - 18; http://dx.doi.org/10.1091/mbc.11.11.4005; PMID: 11071923
- Grzmil M, Rzymski T, Milani M, Harris AL, Capper RG, Saunders NJ, Salhan A, Ragoussis J, Norbury CJ. An oncogenic role of eIF3e/INT6 in human breast cancer. Oncogene 2010; 29:4080 - 9; http://dx.doi.org/10.1038/onc.2010.152; PMID: 20453879
- Rozen F, Edery I, Meerovitch K, Dever TE, Merrick WC, Sonenberg N. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol Cell Biol 1990; 10:1134 - 44; PMID: 2304461
- Shuda M, Kondoh N, Tanaka K, Ryo A, Wakatsuki T, Hada A, Goseki N, Igari T, Hatsuse K, Aihara T, et al. Enhanced expression of translation factor mRNAs in hepatocellular carcinoma. Anticancer Res 2000; 20:2489 - 94; PMID: 10953316
- Eberle J, Krasagakis K, Orfanos CE. Translation initiation factor eIF-4A1 mRNA is consistently overexpressed in human melanoma cells in vitro. Int J Cancer 1997; 71:396 - 401; http://dx.doi.org/10.1002/(SICI)1097-0215(19970502)71:3<396::AID-IJC16>3.0.CO;2-E; PMID: 9139875
- Schroeder JA, Masri AA, Adriance MC, Tessier JC, Kotlarczyk KL, Thompson MC, Gendler SJ. MUC1 overexpression results in mammary gland tumorigenesis and prolonged alveolar differentiation. Oncogene 2004; 23:5739 - 47; http://dx.doi.org/10.1038/sj.onc.1207713; PMID: 15221004
- Kufe DW. Mucins in cancer: function, prognosis and therapy. Nat Rev Cancer 2009; 9:874 - 85; http://dx.doi.org/10.1038/nrc2761; PMID: 19935676
- Jin C, Rajabi H, Rodrigo CM, Porco JA Jr., Kufe D. Targeting the eIF4A RNA helicase blocks translation of the MUC1-C oncoprotein. Oncogene 2013; 32:2179 - 88; http://dx.doi.org/10.1038/onc.2012.236; PMID: 22689062
- Bordeleau ME, Robert F, Gerard B, Lindqvist L, Chen SM, Wendel HG, Brem B, Greger H, Lowe SW, Porco JA Jr., et al. Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model. J Clin Invest 2008; 118:2651 - 60; PMID: 18551192
- Lucas DM, Edwards RB, Lozanski G, West DA, Shin JD, Vargo MA, Davis ME, Rozewski DM, Johnson AJ, Su BN, et al. The novel plant-derived agent silvestrol has B-cell selective activity in chronic lymphocytic leukemia and acute lymphoblastic leukemia in vitro and in vivo. Blood 2009; 113:4656 - 66; http://dx.doi.org/10.1182/blood-2008-09-175430; PMID: 19190247
- Schatz JH, Oricchio E, Wolfe AL, Jiang M, Linkov I, Maragulia J, Shi W, Zhang Z, Rajasekhar VK, Pagano NC, et al. Targeting cap-dependent translation blocks converging survival signals by AKT and PIM kinases in lymphoma. J Exp Med 2011; 208:1799 - 807; http://dx.doi.org/10.1084/jem.20110846; PMID: 21859846
- Waskiewicz AJ, Johnson JC, Penn B, Mahalingam M, Kimball SR, Cooper JA. Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Mol Cell Biol 1999; 19:1871 - 80; PMID: 10022874
- Joshi B, Cai AL, Keiper BD, Minich WB, Mendez R, Beach CM, Stepinski J, Stolarski R, Darzynkiewicz E, Rhoads RE. Phosphorylation of eukaryotic protein synthesis initiation factor 4E at Ser-209. J Biol Chem 1995; 270:14597 - 603; http://dx.doi.org/10.1074/jbc.270.24.14597; PMID: 7782323
- Avdulov S, Li S, Michalek V, Burrichter D, Peterson M, Perlman DM, Manivel JC, Sonenberg N, Yee D, Bitterman PB, et al. Activation of translation complex eIF4F is essential for the genesis and maintenance of the malignant phenotype in human mammary epithelial cells. Cancer Cell 2004; 5:553 - 63; http://dx.doi.org/10.1016/j.ccr.2004.05.024; PMID: 15193258
- De Benedetti A, Graff JR. eIF-4E expression and its role in malignancies and metastases. Oncogene 2004; 23:3189 - 99; http://dx.doi.org/10.1038/sj.onc.1207545; PMID: 15094768
- Zimmer SG, DeBenedetti A, Graff JR. Translational control of malignancy: the mRNA cap-binding protein, eIF-4E, as a central regulator of tumor formation, growth, invasion and metastasis. Anticancer Res 2000; 20:3A 1343 - 51; PMID: 10928042
- Culjkovic B, Tan K, Orolicki S, Amri A, Meloche S, Borden KL. The eIF4E RNA regulon promotes the Akt signaling pathway. J Cell Biol 2008; 181:51 - 63; http://dx.doi.org/10.1083/jcb.200707018; PMID: 18391071
- Li S, Takasu T, Perlman DM, Peterson MS, Burrichter D, Avdulov S, Bitterman PB, Polunovsky VA. Translation factor eIF4E rescues cells from Myc-dependent apoptosis by inhibiting cytochrome c release. J Biol Chem 2003; 278:3015 - 22; http://dx.doi.org/10.1074/jbc.M208821200; PMID: 12441348
- Rhoads RE. Regulation of eukaryotic protein synthesis by initiation factors. J Biol Chem 1993; 268:3017 - 20; PMID: 8428975
- Rosenwald IB, Kaspar R, Rousseau D, Gehrke L, Leboulch P, Chen JJ, Schmidt EV, Sonenberg N, London IM. Eukaryotic translation initiation factor 4E regulates expression of cyclin D1 at transcriptional and post-transcriptional levels. J Biol Chem 1995; 270:21176 - 80; http://dx.doi.org/10.1074/jbc.270.36.21176; PMID: 7673150
- Bitterman PB, Polunovsky VA. Attacking a nexus of the oncogenic circuitry by reversing aberrant eIF4F-mediated translation. Mol Cancer Ther 2012; 11:1051 - 61; http://dx.doi.org/10.1158/1535-7163.MCT-11-0530; PMID: 22572598
- Feoktistova K, Tuvshintogs E, Do A, Fraser CS. Human eIF4E promotes mRNA restructuring by stimulating eIF4A helicase activity. Proc Natl Acad Sci U S A 2013; 110:13339 - 44; http://dx.doi.org/10.1073/pnas.1303781110; PMID: 23901100
- Larsson O, Li S, Issaenko OA, Avdulov S, Peterson M, Smith K, Bitterman PB, Polunovsky VA. Eukaryotic translation initiation factor 4E induced progression of primary human mammary epithelial cells along the cancer pathway is associated with targeted translational deregulation of oncogenic drivers and inhibitors. Cancer Res 2007; 67:6814 - 24; http://dx.doi.org/10.1158/0008-5472.CAN-07-0752; PMID: 17638893
- Ruggero D, Montanaro L, Ma L, Xu W, Londei P, Cordon-Cardo C, Pandolfi PP. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat Med 2004; 10:484 - 6; http://dx.doi.org/10.1038/nm1042; PMID: 15098029
- Zindy P, Bergé Y, Allal B, Filleron T, Pierredon S, Cammas A, Beck S, Mhamdi L, Fan L, Favre G, et al. Formation of the eIF4F translation-initiation complex determines sensitivity to anticancer drugs targeting the EGFR and HER2 receptors. Cancer Res 2011; 71:4068 - 73; http://dx.doi.org/10.1158/0008-5472.CAN-11-0420; PMID: 21498638
- Li BD, Gruner JS, Abreo F, Johnson LW, Yu H, Nawas S, McDonald JC, DeBenedetti A. Prospective study of eukaryotic initiation factor 4E protein elevation and breast cancer outcome. Ann Surg 2002; 235:732 - 8, discussion 738-9; http://dx.doi.org/10.1097/00000658-200205000-00016; PMID: 11981220
- McClusky DR, Chu Q, Yu H, Debenedetti A, Johnson LW, Meschonat C, Turnage R, McDonald JC, Abreo F, Li BD. A prospective trial on initiation factor 4E (eIF4E) overexpression and cancer recurrence in node-positive breast cancer. Ann Surg 2005; 242:584 - 90, discussion 590-2; PMID: 16192819
- Holm N, Byrnes K, Johnson L, Abreo F, Sehon K, Alley J, Meschonat C, Md QC, Li BD. A Prospective Trial on Initiation Factor 4E (eIF4E) Overexpression and Cancer Recurrence in Node-Negative Breast Cancer. Ann Surg Oncol 2008.
- Flowers A, Chu QD, Panu L, Meschonat C, Caldito G, Lowery-Nordberg M, Li BD. Eukaryotic initiation factor 4E overexpression in triple-negative breast cancer predicts a worse outcome. Surgery 2009; 146:220 - 6; http://dx.doi.org/10.1016/j.surg.2009.05.010; PMID: 19628077
- Hiller DJ, Chu Q, Meschonat C, Panu L, Burton G, Li BD. Predictive value of eIF4E reduction after neoadjuvant therapy in breast cancer. J Surg Res 2009; 156:265 - 9; http://dx.doi.org/10.1016/j.jss.2009.03.060; PMID: 19665145
- Graff JR, Konicek BW, Lynch RL, Dumstorf CA, Dowless MS, McNulty AM, Parsons SH, Brail LH, Colligan BM, Koop JW, et al. eIF4E activation is commonly elevated in advanced human prostate cancers and significantly related to reduced patient survival. Cancer Res 2009; 69:3866 - 73; http://dx.doi.org/10.1158/0008-5472.CAN-08-3472; PMID: 19383915
- Coleman LJ, Peter MB, Teall TJ, Brannan RA, Hanby AM, Honarpisheh H, Shaaban AM, Smith L, Speirs V, Verghese ET, et al. Combined analysis of eIF4E and 4E-binding protein expression predicts breast cancer survival and estimates eIF4E activity. Br J Cancer 2009; 100:1393 - 9; http://dx.doi.org/10.1038/sj.bjc.6605044; PMID: 19367274
- Braunstein S, Karpisheva K, Pola C, Goldberg J, Hochman T, Yee H, Cangiarella J, Arju R, Formenti SC, Schneider RJ. A hypoxia-controlled cap-dependent to cap-independent translation switch in breast cancer. Mol Cell 2007; 28:501 - 12; http://dx.doi.org/10.1016/j.molcel.2007.10.019; PMID: 17996713
- Liu J, Stevens PD, Eshleman NE, Gao T. Protein phosphatase PPM1G regulates protein translation and cell growth by dephosphorylating 4E binding protein 1 (4E-BP1). J Biol Chem 2013; 288:23225 - 33; http://dx.doi.org/10.1074/jbc.M113.492371; PMID: 23814053
- Fukuchi-Shimogori T, Ishii I, Kashiwagi K, Mashiba H, Ekimoto H, Igarashi K. Malignant transformation by overproduction of translation initiation factor eIF4G. Cancer Res 1997; 57:5041 - 4; PMID: 9371500
- Ramírez-Valle F, Braunstein S, Zavadil J, Formenti SC, Schneider RJ. eIF4GI links nutrient sensing by mTOR to cell proliferation and inhibition of autophagy. J Cell Biol 2008; 181:293 - 307; http://dx.doi.org/10.1083/jcb.200710215; PMID: 18426977
- Silvera D, Arju R, Darvishian F, Levine PH, Zolfaghari L, Goldberg J, Hochman T, Formenti SC, Schneider RJ. Essential role for eIF4GI overexpression in the pathogenesis of inflammatory breast cancer. Nat Cell Biol 2009; 11:903 - 8; http://dx.doi.org/10.1038/ncb1900; PMID: 19525934
- Mahoney SJ, Dempsey JM, Blenis J. Cell signaling in protein synthesis ribosome biogenesis and translation initiation and elongation. Prog Mol Biol Transl Sci 2009; 90:53 - 107; http://dx.doi.org/10.1016/S1877-1173(09)90002-3; PMID: 20374739
- Wu H, Yang JM, Jin S, Zhang H, Hait WN. Elongation factor-2 kinase regulates autophagy in human glioblastoma cells. Cancer Res 2006; 66:3015 - 23; http://dx.doi.org/10.1158/0008-5472.CAN-05-1554; PMID: 16540650
- Arora S, Yang JM, Kinzy TG, Utsumi R, Okamoto T, Kitayama T, Ortiz PA, Hait WN. Identification and characterization of an inhibitor of eukaryotic elongation factor 2 kinase against human cancer cell lines. Cancer Res 2003; 63:6894 - 9; PMID: 14583488
- Alaiya AA, Franzén B, Fujioka K, Moberger B, Schedvins K, Silfversvärd C, Linder S, Auer G. Phenotypic analysis of ovarian carcinoma: polypeptide expression in benign, borderline and malignant tumors. Int J Cancer 1997; 73:678 - 83; http://dx.doi.org/10.1002/(SICI)1097-0215(19971127)73:5<678::AID-IJC11>3.0.CO;2-2; PMID: 9398045
- Nakamura J, Aoyagi S, Nanchi I, Nakatsuka S, Hirata E, Shibata S, Fukuda M, Yamamoto Y, Fukuda I, Tatsumi N, et al. Overexpression of eukaryotic elongation factor eEF2 in gastrointestinal cancers and its involvement in G2/M progression in the cell cycle. Int J Oncol 2009; 34:1181 - 9; PMID: 19360331
- Cheng Y, Ren X, Zhang Y, Patel R, Sharma A, Wu H, Robertson GP, Yan L, Rubin E, Yang JM. eEF-2 kinase dictates cross-talk between autophagy and apoptosis induced by Akt Inhibition, thereby modulating cytotoxicity of novel Akt inhibitor MK-2206. Cancer Res 2011; 71:2654 - 63; http://dx.doi.org/10.1158/0008-5472.CAN-10-2889; PMID: 21307130
- Bagaglio DM, Cheng EH, Gorelick FS, Mitsui K, Nairn AC, Hait WN. Phosphorylation of elongation factor 2 in normal and malignant rat glial cells. Cancer Res 1993; 53:Suppl 2260 - 4; PMID: 8485712
- Arora S, Yang JM, Hait WN. Identification of the ubiquitin-proteasome pathway in the regulation of the stability of eukaryotic elongation factor-2 kinase. Cancer Res 2005; 65:3806 - 10; http://dx.doi.org/10.1158/0008-5472.CAN-04-4036; PMID: 15867377
- Cheng EH, Gorelick FS, Czernik AJ, Bagaglio DM, Hait WN. Calmodulin-dependent protein kinases in rat glioblastoma. Cell Growth Differ 1995; 6:615 - 21; PMID: 7647041
- Parmer TG, Ward MD, Yurkow EJ, Vyas VH, Kearney TJ, Hait WN. Activity and regulation by growth factors of calmodulin-dependent protein kinase III (elongation factor 2-kinase) in human breast cancer. Br J Cancer 1999; 79:59 - 64; http://dx.doi.org/10.1038/sj.bjc.6690012; PMID: 10408694
- Tekedereli I, Alpay SN, Tavares CD, Cobanoglu ZE, Kaoud TS, Sahin I, Sood AK, Lopez-Berestein G, Dalby KN, Ozpolat B. Targeted silencing of elongation factor 2 kinase suppresses growth and sensitizes tumors to doxorubicin in an orthotopic model of breast cancer. PLoS One 2012; 7:e41171; http://dx.doi.org/10.1371/journal.pone.0041171; PMID: 22911754
- Wen YH, Shi X, Chiriboga L, Matsahashi S, Yee H, Afonja O. Alterations in the expression of PDCD4 in ductal carcinoma of the breast. Oncol Rep 2007; 18:1387 - 93; PMID: 17982621
- Yang HS, Jansen AP, Nair R, Shibahara K, Verma AK, Cmarik JL, Colburn NH. A novel transformation suppressor, Pdcd4, inhibits AP-1 transactivation but not NF-kappaB or ODC transactivation. Oncogene 2001; 20:669 - 76; http://dx.doi.org/10.1038/sj.onc.1204137; PMID: 11314000
- Yang HS, Jansen AP, Komar AA, Zheng X, Merrick WC, Costes S, Lockett SJ, Sonenberg N, Colburn NH. The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol Cell Biol 2003; 23:26 - 37; http://dx.doi.org/10.1128/MCB.23.1.26-37.2003; PMID: 12482958
- Schmid T, Jansen AP, Baker AR, Hegamyer G, Hagan JP, Colburn NH. Translation inhibitor Pdcd4 is targeted for degradation during tumor promotion. Cancer Res 2008; 68:1254 - 60; http://dx.doi.org/10.1158/0008-5472.CAN-07-1719; PMID: 18296647
- Teske BF, Baird TD, Wek RC. Methods for analyzing eIF2 kinases and translational control in the unfolded protein response. Methods Enzymol 2011; 490:333 - 56; http://dx.doi.org/10.1016/B978-0-12-385114-7.00019-2; PMID: 21266259
- Backer MV, Backer JM, Chinnaiyan P. Targeting the unfolded protein response in cancer therapy. Methods Enzymol 2011; 491:37 - 56; http://dx.doi.org/10.1016/B978-0-12-385928-0.00003-1; PMID: 21329793
- Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol 2012; 6:590 - 610; http://dx.doi.org/10.1016/j.molonc.2012.09.006; PMID: 23102669
- Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science 2005; 309:1573 - 6; http://dx.doi.org/10.1126/science.1115079; PMID: 16081698
- Kentsis A, Topisirovic I, Culjkovic B, Shao L, Borden KL. Ribavirin suppresses eIF4E-mediated oncogenic transformation by physical mimicry of the 7-methyl guanosine mRNA cap. Proc Natl Acad Sci U S A 2004; 101:18105 - 10; http://dx.doi.org/10.1073/pnas.0406927102; PMID: 15601771
- Soni A, Akcakanat A, Singh G, Luyimbazi D, Zheng Y, Kim D, Gonzalez-Angulo A, Meric-Bernstam F. eIF4E knockdown decreases breast cancer cell growth without activating Akt signaling. Mol Cancer Ther 2008; 7:1782 - 8; http://dx.doi.org/10.1158/1535-7163.MCT-07-2357; PMID: 18644990
- Graff JR, Konicek BW, Vincent TM, Lynch RL, Monteith D, Weir SN, Schwier P, Capen A, Goode RL, Dowless MS, et al. Therapeutic suppression of translation initiation factor eIF4E expression reduces tumor growth without toxicity. J Clin Invest 2007; 117:2638 - 48; http://dx.doi.org/10.1172/JCI32044; PMID: 17786246
- Hong DS, Kurzrock R, Oh Y, Wheler J, Naing A, Brail L, Callies S, André V, Kadam SK, Nasir A, et al. A phase 1 dose escalation, pharmacokinetic, and pharmacodynamic evaluation of eIF-4E antisense oligonucleotide LY2275796 in patients with advanced cancer. Clin Cancer Res 2011; 17:6582 - 91; http://dx.doi.org/10.1158/1078-0432.CCR-11-0430; PMID: 21831956
- Pharmaceuticals I. Safety and Tolerability Study of ISIS EIF4E Rx in Combination With Carboplatin and Paclitaxel (NSCLC). In: ClinicalTrials.gov [Internet]. Bethesda (MD): National Library of Medicine (US). 2010- [cited 2014 March 27]. Available from: http://clinicaltrials.gov/ct2/show/NCT01234038 NLM identifier: NCT1234038.
- Pharmaceuticals I. Safety and Tolerability Study of ISIS EIF4E Rx in Combination With Docetaxel and Prednisone (CRPC). In: ClinicalTrials.gov [Internet]. Bethesda (MD): National Library of Medicine (US). 2010- [cited 2014 March 27]. Available from: http://clinicaltrials.gov/ct2/show/NCT01234025 NLM identifier: NCT01234025.
- Ghosh B, Benyumov AO, Ghosh P, Jia Y, Avdulov S, Dahlberg PS, Peterson M, Smith K, Polunovsky VA, Bitterman PB, et al. Nontoxic chemical interdiction of the epithelial-to-mesenchymal transition by targeting cap-dependent translation. ACS Chem Biol 2009; 4:367 - 77; http://dx.doi.org/10.1021/cb9000475; PMID: 19351181
- Assouline S, Culjkovic B, Cocolakis E, Rousseau C, Beslu N, Amri A, Caplan S, Leber B, Roy DC, Miller WH Jr., et al. Molecular targeting of the oncogene eIF4E in acute myeloid leukemia (AML): a proof-of-principle clinical trial with ribavirin. Blood 2009; 114:257 - 60; http://dx.doi.org/10.1182/blood-2009-02-205153; PMID: 19433856
- Borden KL. Targeting the oncogene eIF4E in cancer: From the bench to clinical trials. Clin Invest Med 2011; 34:E315; PMID: 22129918
- Kraljacic BC, Arguello M, Amri A, Cormack G, Borden K. Inhibition of eIF4E with ribavirin cooperates with common chemotherapies in primary acute myeloid leukemia specimens. Leukemia 2011; 25:1197 - 200; http://dx.doi.org/10.1038/leu.2011.57; PMID: 21455212
- Jewish General Hospital. A Phase I/II Study of Ribavirin and Low-dose Cytarabine Arabinoside (Ara-C) in Acute Myeloid Leukemia (AML) M4 and M5 Subtypes, and AML With High eIF4E Expression. In: ClinicalTrials.gov [Internet]. Bethesda (MD): National Library of Medicine (US). 2010- [cited 2014 March 27]. Available from: http://clinicaltrials.gov/ct2/show/record/NCT01056523 NLM identifier: NCT01056523.
- Jewish General Hospital. A Phase I/II Exploratory Study of Ribavirin in Metastatic Breast Cancer Expressing Elevated eIF4E. In: ClinicalTrials.gov [Internet]. Bethesda (MD): National Library of Medicine (US). 2010- [cited 2014 March 27]. Available from: http://clinicaltrials.gov/ct2/show/NCT01056757 NLM identifier: NCT01056757.
- Westman B, Beeren L, Grudzien E, Stepinski J, Worch R, Zuberek J, Jemielity J, Stolarski R, Darzynkiewicz E, Rhoads RE, et al. The antiviral drug ribavirin does not mimic the 7-methylguanosine moiety of the mRNA cap structure in vitro. RNA 2005; 11:1505 - 13; http://dx.doi.org/10.1261/rna.2132505; PMID: 16131589
- Yan Y, Svitkin Y, Lee JM, Bisaillon M, Pelletier J. Ribavirin is not a functional mimic of the 7-methyl guanosine mRNA cap. RNA 2005; 11:1238 - 44; http://dx.doi.org/10.1261/rna.2930805; PMID: 16043507
- Herbert TP, Fåhraeus R, Prescott A, Lane DP, Proud CG. Rapid induction of apoptosis mediated by peptides that bind initiation factor eIF4E. Curr Biol 2000; 10:793 - 6; http://dx.doi.org/10.1016/S0960-9822(00)00567-4; PMID: 10898981
- Salaün P, Boulben S, Mulner-Lorillon O, Bellé R, Sonenberg N, Morales J, Cormier P. Embryonic-stage-dependent changes in the level of eIF4E-binding proteins during early development of sea urchin embryos. J Cell Sci 2005; 118:1385 - 94; http://dx.doi.org/10.1242/jcs.01716; PMID: 15769855
- Ko SY, Guo H, Barengo N, Naora H. Inhibition of ovarian cancer growth by a tumor-targeting peptide that binds eukaryotic translation initiation factor 4E. Clin Cancer Res 2009; 15:4336 - 47; http://dx.doi.org/10.1158/1078-0432.CCR-08-2924; PMID: 19458052
- Moerke NJ, Aktas H, Chen H, Cantel S, Reibarkh MY, Fahmy A, Gross JD, Degterev A, Yuan J, Chorev M, et al. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell 2007; 128:257 - 67; http://dx.doi.org/10.1016/j.cell.2006.11.046; PMID: 17254965
- Cencic R, Hall DR, Robert F, Du Y, Min J, Li L, Qui M, Lewis I, Kurtkaya S, Dingledine R, et al. Reversing chemoresistance by small molecule inhibition of the translation initiation complex eIF4F. Proc Natl Acad Sci U S A 2011; 108:1046 - 51; http://dx.doi.org/10.1073/pnas.1011477108; PMID: 21191102
- Bordeleau ME, Matthews J, Wojnar JM, Lindqvist L, Novac O, Jankowsky E, Sonenberg N, Northcote P, Teesdale-Spittle P, Pelletier J. Stimulation of mammalian translation initiation factor eIF4A activity by a small molecule inhibitor of eukaryotic translation. Proc Natl Acad Sci U S A 2005; 102:10460 - 5; http://dx.doi.org/10.1073/pnas.0504249102; PMID: 16030146
- Bordeleau ME, Cencic R, Lindqvist L, Oberer M, Northcote P, Wagner G, Pelletier J. RNA-mediated sequestration of the RNA helicase eIF4A by Pateamine A inhibits translation initiation. Chem Biol 2006; 13:1287 - 95; http://dx.doi.org/10.1016/j.chembiol.2006.10.005; PMID: 17185224
- Kuznetsov G, Xu Q, Rudolph-Owen L, Tendyke K, Liu J, Towle M, Zhao N, Marsh J, Agoulnik S, Twine N, et al. Potent in vitro and in vivo anticancer activities of des-methyl, des-amino pateamine A, a synthetic analogue of marine natural product pateamine A. Mol Cancer Ther 2009; 8:1250 - 60; http://dx.doi.org/10.1158/1535-7163.MCT-08-1026; PMID: 19417157
- Cencic R, Carrier M, Galicia-Vázquez G, Bordeleau ME, Sukarieh R, Bourdeau A, Brem B, Teodoro JG, Greger H, Tremblay ML, et al. Antitumor activity and mechanism of action of the cyclopenta[b]benzofuran, silvestrol. PLoS One 2009; 4:e5223; http://dx.doi.org/10.1371/journal.pone.0005223; PMID: 19401772
- Bordeleau ME, Mori A, Oberer M, Lindqvist L, Chard LS, Higa T, Belsham GJ, Wagner G, Tanaka J, Pelletier J. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat Chem Biol 2006; 2:213 - 20; http://dx.doi.org/10.1038/nchembio776; PMID: 16532013
- Tsumuraya T, Ishikawa C, Machijima Y, Nakachi S, Senba M, Tanaka J, Mori N. Effects of hippuristanol, an inhibitor of eIF4A, on adult T-cell leukemia. Biochem Pharmacol 2011; 81:713 - 22; http://dx.doi.org/10.1016/j.bcp.2010.12.025; PMID: 21219881
- Shi XP, Yin KC, Ahern J, Davis LJ, Stern AM, Waxman L. Effects of N1-guanyl-1,7-diaminoheptane, an inhibitor of deoxyhypusine synthase, on the growth of tumorigenic cell lines in culture. Biochim Biophys Acta 1996; 1310:119 - 26; http://dx.doi.org/10.1016/0167-4889(95)00165-4; PMID: 9244184
- Clement PM, Hanauske-Abel HM, Wolff EC, Kleinman HK, Park MH. The antifungal drug ciclopirox inhibits deoxyhypusine and proline hydroxylation, endothelial cell growth and angiogenesis in vitro. Int J Cancer 2002; 100:491 - 8; http://dx.doi.org/10.1002/ijc.10515; PMID: 12115536
- Zhou H, Shen T, Luo Y, Liu L, Chen W, Xu B, Han X, Pang J, Rivera CA, Huang S. The antitumor activity of the fungicide ciclopirox. Int J Cancer 2010; 127:2467 - 77; http://dx.doi.org/10.1002/ijc.25255; PMID: 20225320
- Jasiulionis MG, Luchessi AD, Moreira AG, Souza PP, Suenaga AP, Correa M, Costa CA, Curi R, Costa-Neto CM. Inhibition of eukaryotic translation initiation factor 5A (eIF5A) hypusination impairs melanoma growth. Cell Biochem Funct 2007; 25:109 - 14; http://dx.doi.org/10.1002/cbf.1351; PMID: 16850525
- Eberhard Y, McDermott SP, Wang X, Gronda M, Venugopal A, Wood TE, Hurren R, Datti A, Batey RA, Wrana J, et al. Chelation of intracellular iron with the antifungal agent ciclopirox olamine induces cell death in leukemia and myeloma cells. Blood 2009; 114:3064 - 73; http://dx.doi.org/10.1182/blood-2009-03-209965; PMID: 19589922
- Redpath NT, Foulstone EJ, Proud CG. Regulation of translation elongation factor-2 by insulin via a rapamycin-sensitive signalling pathway. EMBO J 1996; 15:2291 - 7; PMID: 8641294
- Shor B, Zhang WG, Toral-Barza L, Lucas J, Abraham RT, Gibbons JJ, Yu K. A new pharmacologic action of CCI-779 involves FKBP12-independent inhibition of mTOR kinase activity and profound repression of global protein synthesis. Cancer Res 2008; 68:2934 - 43; http://dx.doi.org/10.1158/0008-5472.CAN-07-6487; PMID: 18413763
- Huang MT. Harringtonine, an inhibitor of initiation of protein biosynthesis. Mol Pharmacol 1975; 11:511 - 9; PMID: 1237080
- Fresno M, Jiménez A, Vázquez D. Inhibition of translation in eukaryotic systems by harringtonine. Eur J Biochem 1977; 72:323 - 30; http://dx.doi.org/10.1111/j.1432-1033.1977.tb11256.x; PMID: 319998
- Gürel G, Blaha G, Moore PB, Steitz TA. U2504 determines the species specificity of the A-site cleft antibiotics: the structures of tiamulin, homoharringtonine, and bruceantin bound to the ribosome. J Mol Biol 2009; 389:146 - 56; http://dx.doi.org/10.1016/j.jmb.2009.04.005; PMID: 19362093
- Wetzler M, Segal D. Omacetaxine as an anticancer therapeutic: what is old is new again. Curr Pharm Des 2011; 17:59 - 64; http://dx.doi.org/10.2174/138161211795049778; PMID: 21294709
- Memmott RM, Dennis PA. Akt-dependent and -independent mechanisms of mTOR regulation in cancer. Cell Signal 2009; 21:656 - 64; http://dx.doi.org/10.1016/j.cellsig.2009.01.004; PMID: 19166931
- Mondesire WH, Jian W, Zhang H, Ensor J, Hung MC, Mills GB, Meric-Bernstam F. Targeting mammalian target of rapamycin synergistically enhances chemotherapy-induced cytotoxicity in breast cancer cells. Clin Cancer Res 2004; 10:7031 - 42; http://dx.doi.org/10.1158/1078-0432.CCR-04-0361; PMID: 15501983
- Hudes G, Carducci M, Tomczak P, Dutcher J, Figlin R, Kapoor A, Staroslawska E, Sosman J, McDermott D, Bodrogi I, et al, Global ARCC Trial. Temsirolimus, interferon alfa, or both for advanced renal-cell carcinoma. N Engl J Med 2007; 356:2271 - 81; http://dx.doi.org/10.1056/NEJMoa066838; PMID: 17538086
- Motzer RJ, Escudier B, Oudard S, Hutson TE, Porta C, Bracarda S, Grünwald V, Thompson JA, Figlin RA, Hollaender N, et al, RECORD-1 Study Group. Efficacy of everolimus in advanced renal cell carcinoma: a double-blind, randomised, placebo-controlled phase III trial. Lancet 2008; 372:449 - 56; http://dx.doi.org/10.1016/S0140-6736(08)61039-9; PMID: 18653228
- Yao JC, Shah MH, Ito T, Bohas CL, Wolin EM, Van Cutsem E, Hobday TJ, Okusaka T, Capdevila J, de Vries EG, et al, RAD001 in Advanced Neuroendocrine Tumors, Third Trial (RADIANT-3) Study Group. Everolimus for advanced pancreatic neuroendocrine tumors. N Engl J Med 2011; 364:514 - 23; http://dx.doi.org/10.1056/NEJMoa1009290; PMID: 21306238
- Franz DN, Belousova E, Sparagana S, Bebin EM, Frost M, Kuperman R, Witt O, Kohrman MH, Flamini JR, Wu JY, et al. Efficacy and safety of everolimus for subependymal giant cell astrocytomas associated with tuberous sclerosis complex (EXIST-1): a multicentre, randomised, placebo-controlled phase 3 trial. Lancet 2013; 381:125 - 32; http://dx.doi.org/10.1016/S0140-6736(12)61134-9; PMID: 23158522
- Bissler JJ, Kingswood JC, Radzikowska E, Zonnenberg BA, Frost M, Belousova E, Sauter M, Nonomura N, Brakemeier S, de Vries PJ, et al. Everolimus for angiomyolipoma associated with tuberous sclerosis complex or sporadic lymphangioleiomyomatosis (EXIST-2): a multicentre, randomised, double-blind, placebo-controlled trial. Lancet 2013; 381:817 - 24; http://dx.doi.org/10.1016/S0140-6736(12)61767-X; PMID: 23312829
- Ellard SL, Clemons M, Gelmon KA, Norris B, Kennecke H, Chia S, Pritchard K, Eisen A, Vandenberg T, Taylor M, et al. Randomized phase II study comparing two schedules of everolimus in patients with recurrent/metastatic breast cancer: NCIC Clinical Trials Group IND.163. J Clin Oncol 2009; 27:4536 - 41; http://dx.doi.org/10.1200/JCO.2008.21.3033; PMID: 19687332
- Boulay A, Rudloff J, Ye J, Zumstein-Mecker S, O’Reilly T, Evans DB, Chen S, Lane HA. Dual inhibition of mTOR and estrogen receptor signaling in vitro induces cell death in models of breast cancer. Clin Cancer Res 2005; 11:5319 - 28; http://dx.doi.org/10.1158/1078-0432.CCR-04-2402; PMID: 16033851
- Bachelot T, Bourgier C, Cropet C, Ray-Coquard I, Ferrero JM, Freyer G, Abadie-Lacourtoisie S, Eymard JC, Debled M, Spaëth D, et al. Randomized phase II trial of everolimus in combination with tamoxifen in patients with hormone receptor-positive, human epidermal growth factor receptor 2-negative metastatic breast cancer with prior exposure to aromatase inhibitors: a GINECO study. J Clin Oncol 2012; 30:2718 - 24; http://dx.doi.org/10.1200/JCO.2011.39.0708; PMID: 22565002
- Baselga J, Semiglazov V, van Dam P, Manikhas A, Bellet M, Mayordomo J, Campone M, Kubista E, Greil R, Bianchi G, et al. Phase II randomized study of neoadjuvant everolimus plus letrozole compared with placebo plus letrozole in patients with estrogen receptor-positive breast cancer. J Clin Oncol 2009; 27:2630 - 7; http://dx.doi.org/10.1200/JCO.2008.18.8391; PMID: 19380449
- Baselga J, Campone M, Piccart M, Burris HA 3rd, Rugo HS, Sahmoud T, Noguchi S, Gnant M, Pritchard KI, Lebrun F, et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med 2012; 366:520 - 9; http://dx.doi.org/10.1056/NEJMoa1109653; PMID: 22149876
- Beaver JA, Park BH. The BOLERO-2 trial: the addition of everolimus to exemestane in the treatment of postmenopausal hormone receptor-positive advanced breast cancer. Future Oncol 2012; 8:651 - 7; http://dx.doi.org/10.2217/fon.12.49; PMID: 22764762
- Wolff AC, Lazar AA, Bondarenko I, Garin AM, Brincat S, Chow L, Sun Y, Neskovic-Konstantinovic Z, Guimaraes RC, Fumoleau P, et al. Randomized phase III placebo-controlled trial of letrozole plus oral temsirolimus as first-line endocrine therapy in postmenopausal women with locally advanced or metastatic breast cancer. J Clin Oncol 2013; 31:195 - 202; http://dx.doi.org/10.1200/JCO.2011.38.3331; PMID: 23233719
- Mazzoletti M, Bortolin F, Brunelli L, Pastorelli R, Di Giandomenico S, Erba E, Ubezio P, Broggini M. Combination of PI3K/mTOR inhibitors: antitumor activity and molecular correlates. Cancer Res 2011; 71:4573 - 84; http://dx.doi.org/10.1158/0008-5472.CAN-10-4322; PMID: 21602434
- Li T, Wang J, Wang X, Yang N, Chen SM, Tong LJ, Yang CH, Meng LH, Ding J. WJD008, a dual phosphatidylinositol 3-kinase (PI3K)/mammalian target of rapamycin inhibitor, prevents PI3K signaling and inhibits the proliferation of transformed cells with oncogenic PI3K mutant. J Pharmacol Exp Ther 2010; 334:830 - 8; http://dx.doi.org/10.1124/jpet.110.167940; PMID: 20522531
- Serra V, Markman B, Scaltriti M, Eichhorn PJ, Valero V, Guzman M, Botero ML, Llonch E, Atzori F, Di Cosimo S, et al. NVP-BEZ235, a dual PI3K/mTOR inhibitor, prevents PI3K signaling and inhibits the growth of cancer cells with activating PI3K mutations. Cancer Res 2008; 68:8022 - 30; http://dx.doi.org/10.1158/0008-5472.CAN-08-1385; PMID: 18829560
- Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol 2002; 4:648 - 57; http://dx.doi.org/10.1038/ncb839; PMID: 12172553
- National Cancer Institute. Phase II Trial of Akt Inhibitor MK2206 in Patients With Advanced Breast Cancer Who Have Tumors With a PIK3CA Mutation, or an AKT Mutation, and/or PTEN Loss/PTEN Mutation. In: ClinicalTrials.gov [Internet]. Bethesda (MD): National Library of Medicine (US). 2011- [cited 2014 March 27]. Available from: http://clinicaltrials.gov/ct2/show/NCT01277757 NLM identifier: NCT01277757.
- QuantumLeap Healthcare Collaborative. I-SPY 2 Trial (Investigation of Serial Studies to Predict Your Therapeutic Response With Imaging And moLecular Analysis 2). In: ClinicalTrials.gov [Internet]. Bethesda (MD): National Library of Medicine (US). 2009- [cited 2014 March 27]. Available from: http://clinicaltrials.gov/ct2/show/NCT01042379 NLM identifier: NCT01042379.
- Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987; 235:177 - 82; http://dx.doi.org/10.1126/science.3798106; PMID: 3798106
- Carracedo A, Ma L, Teruya-Feldstein J, Rojo F, Salmena L, Alimonti A, Egia A, Sasaki AT, Thomas G, Kozma SC, et al. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J Clin Invest 2008; 118:3065 - 74; PMID: 18725988
- Carrière A, Cargnello M, Julien LA, Gao H, Bonneil E, Thibault P, Roux PP. Oncogenic MAPK signaling stimulates mTORC1 activity by promoting RSK-mediated raptor phosphorylation. Curr Biol 2008; 18:1269 - 77; http://dx.doi.org/10.1016/j.cub.2008.07.078; PMID: 18722121
- Carriere A, Romeo Y, Acosta-Jaquez HA, Moreau J, Bonneil E, Thibault P, Fingar DC, Roux PP. ERK1/2 phosphorylate Raptor to promote Ras-dependent activation of mTOR complex 1 (mTORC1). J Biol Chem 2011; 286:567 - 77; http://dx.doi.org/10.1074/jbc.M110.159046; PMID: 21071439
- Wheater MJ, Johnson PW, Blaydes JP. The role of MNK proteins and eIF4E phosphorylation in breast cancer cell proliferation and survival. Cancer Biol Ther 2010; 10:728 - 35; http://dx.doi.org/10.4161/cbt.10.7.12965; PMID: 20686366
- Konicek BW, Stephens JR, McNulty AM, Robichaud N, Peery RB, Dumstorf CA, Dowless MS, Iversen PW, Parsons S, Ellis KE, et al. Therapeutic inhibition of MAP kinase interacting kinase blocks eukaryotic initiation factor 4E phosphorylation and suppresses outgrowth of experimental lung metastases. Cancer Res 2011; 71:1849 - 57; http://dx.doi.org/10.1158/0008-5472.CAN-10-3298; PMID: 21233335
- Hvidtfeldt UA, Gunter MJ, Lange T, Chlebowski RT, Lane D, Farhat GN, Freiberg MS, Keiding N, Lee JS, Prentice R, et al. Quantifying mediating effects of endogenous estrogen and insulin in the relation between obesity, alcohol consumption, and breast cancer. Cancer Epidemiol Biomarkers Prev 2012; 21:1203 - 12; http://dx.doi.org/10.1158/1055-9965.EPI-12-0310; PMID: 22564867
- Jiralerspong S, Palla SL, Giordano SH, Meric-Bernstam F, Liedtke C, Barnett CM, Hsu L, Hung MC, Hortobagyi GN, Gonzalez-Angulo AM. Metformin and pathologic complete responses to neoadjuvant chemotherapy in diabetic patients with breast cancer. J Clin Oncol 2009; 27:3297 - 302; http://dx.doi.org/10.1200/JCO.2009.19.6410; PMID: 19487376
- Alimova IN, Liu B, Fan Z, Edgerton SM, Dillon T, Lind SE, Thor AD. Metformin inhibits breast cancer cell growth, colony formation and induces cell cycle arrest in vitro. Cell Cycle 2009; 8:909 - 15; http://dx.doi.org/10.4161/cc.8.6.7933; PMID: 19221498
- Liu H, Scholz C, Zang C, Schefe JH, Habbel P, Regierer AC, Schulz CO, Possinger K, Eucker J. Metformin and the mTOR inhibitor everolimus (RAD001) sensitize breast cancer cells to the cytotoxic effect of chemotherapeutic drugs in vitro. Anticancer Res 2012; 32:1627 - 37; PMID: 22593441
- Col NF, Ochs L, Springmann V, Aragaki AK, Chlebowski RT. Metformin and breast cancer risk: a meta-analysis and critical literature review. Breast Cancer Res Treat 2012; 135:639 - 46; http://dx.doi.org/10.1007/s10549-012-2170-x; PMID: 22847511
- Niraula S, Dowling RJ, Ennis M, Chang MC, Done SJ, Hood N, Escallon J, Leong WL, McCready DR, Reedijk M, et al. Metformin in early breast cancer: a prospective window of opportunity neoadjuvant study. Breast Cancer Res Treat 2012; 135:821 - 30; http://dx.doi.org/10.1007/s10549-012-2223-1; PMID: 22933030
- Chen Y, Li S. Omacetaxine mepesuccinate in the treatment of intractable chronic myeloid leukemia. Onco Targets Ther 2014; 7:177 - 86; PMID: 24516334
- Cortes JE, Nicolini FE, Wetzler M, Lipton JH, Akard L, Craig A, Nanda N, Benichou AC, Leonoudakis J, Khoury HJ, et al. Subcutaneous omacetaxine mepesuccinate in patients with chronic-phase chronic myeloid leukemia previously treated with 2 or more tyrosine kinase inhibitors including imatinib. Clin Lymphoma Myeloma Leuk 2013; 13:584 - 91; http://dx.doi.org/10.1016/j.clml.2013.03.020; PMID: 23787123
