Abstract
Replication and transcription machineries progress at high speed on the same DNA template, which inevitably causes traffic accidents. Problems are not only caused by frontal collisions between polymerases, but also by cotranscriptional R-loops. These RNA-DNA hybrids induce genomic instability by blocking fork progression and could be implicated in the development of cancer.
Introduction
DNA replication and transcription are fundamental genetic processes that are essential for cell growth and division. They are carried out by large protein complexes progressing at high speed and for long distances along the chromosomes. Head-on collisions inevitably arise when replisomes and RNA polymerases (RNAP) face each other on the same DNA template. A large body of evidence from both prokaryotes and eukaryotes indicates that frontal collisions induce replication fork stalling and genomic instability.Citation1–Citation7 In E. coli, the replisome moves 15 to 30 times faster than transcription complexes and the replication machinery can also rear-end RNA polymerases.Citation8,Citation9 In general, the replisome is given priority and RNAP are displaced. However, cells lacking both RNAP modulators and fork recovery factors have much reduced viability, indicating that collisions are frequent and have deleterious consequences.Citation10 To limit frontal collisions, bacterial genomes show a bias for genes to be encoded on the leading strand, resulting in coorientation of transcription and replication.Citation11 Reversing this collinear organization in Bacillus subtilis reduces the rate of DNA synthesis by ∼50% specifically in regions with reversed bias.Citation12 Altogether, these data indicate that bacteria have evolved multiple means to minimize interference between replication and transcription.Citation10 Whether the same mechanisms operate in eukaryotic cells is currently unclear. In this review, we focus on the nature of replication/transcription interference in eukaryotes and on its consequences on genome integrity.
Functional Links between Transcription, Replication and Recombination
Although it is now well established that transcription increases genomic instability in eukaryotes,Citation13–Citation15 the molecular mechanisms involved remain poorly understood. Evidence from budding yeast indicates that, like in bacteria, gene expression induces replication fork pausing in eukaryotic cells.Citation7,Citation16–Citation19 Since arrested forks are prone to recombination,Citation20 it has been proposed that transcription induces genomic instability by blocking fork progression.Citation6 This view is supported by yeast studies showing that transcription, replication and recombination are functionally linked.Citation15 Similarly, transcription-associated recombination (TAR) is only detected during S phase in mammalian cells.Citation21 Moreover, TAR is associated with signatures of one-ended double-strand break (DSB) recombination, and not with classical two-ended DSB repair, suggesting that recombination occurs at broken replication forks.Citation21,Citation22 Non-exclusive models have been proposed to explain how transcription interferes with DNA replication in eukaryotes. These models are discussed in the following sections.
Collision between Replication and Transcription Machineries
Several lines of evidence suggest that TAR is promoted by frontal collisions between replication and transcription complexes in eukaryotes, as it is the case in bacteria. These collisions would in turn provoke fork collapse or reversion, which would be resolved by recombinational repair (). In agreement with this view, transcription by RNA polymerase II and III induces fork arrest in S. cerevisiae only when it opposes the direction of replication.Citation7,Citation17 Like bacteria, eukaryotes have also evolved polar replication fork barriers (RFBs) to prevent head-on collisions between polymerases.Citation20 The best-characterized RFB is located downstream of 35S rRNA genes in budding yeast (). This barrier blocks replication forks progressing opposite to the direction of transcriptionCitation23,Citation24 and inactivation of RFBs increases collisions between forks and highly-expressed rRNA genes.Citation18 Mammalian genomes also contain rDNA RFBsCitation25 and their genes are potentially organized collinearly with replication.Citation26 Altogether, these data indicate that eukaryotes have evolved systems to prevent collisions between transcription and replication complexes.
However, other observations also argue against the collision model and indicate that additional mechanisms contribute to replication/transcription interference in eukaryotic cells. Indeed, unlike in bacteria, replication and transcription occur within spatially and temporally separated domains in the eukaryotes.Citation27,Citation28 Moreover, it is generally believed that DNA and RNA polymerases do not travel along the chromosomes but rather form large replication and transcription factories through which DNA is pulled.Citation29,Citation30 Inactive or blocked RNAPs could remain attached to DNA and interfere with replication. Alternatively, replication-impeding structures could form behind RNAPs and persist on DNA after transcription has ceased. These include RNA-DNA hybrids called R-loops, which were shown to affect genomic integrity in eukaryotes and prokaryotes.Citation31–Citation34 Recent data suggesting that R-loops interfere with replication forks are discussed in the following section.
R-Loops Impede Replication Fork Progression
Cotranscriptional R-loops form during transcription when the nascent RNA reanneals with the template DNA strand, leaving the other strand unpaired (). R-loop formation occurs preferentially at GC-rich regions and is favored by negative DNA supercoiling accumulating behind the RNAP.Citation31 Eukaryotic transcription is coupled with multiple processes such as pre-mRNA maturation, surveillance and export. Defects in any of these processes have been implicated in the formation of R-loops.Citation2,Citation6,Citation35 This is best illustrated with the yeast THO/TREX complex, mutation in which leads to defects in transcription elongation and induces TAR.Citation15,Citation34 Importantly, fork progression is impaired at actively transcribed genes in THO mutants and induces a constitutive activation of the replication stress response.Citation36 Overexpression of RNase H, an enzyme that degrades R-loops, suppresses fork arrest and TAR in THO mutants.Citation15 Moreover, one particular THO/TREX mutant showing transcription and RNA export defects but no increased replication fork pausing does not have increased TAR.Citation37 Altogether, these data indicate that replication fork stalling induced by R-loops promote recombinational repair and increase TAR in yeast THO mutants.Citation36
In vertebrates, R-loops also form in cells deficient in the splicing factor ASF/SF2 and induce a type of genomic instability that is suppressed by RNase H.Citation32,Citation35 Other studies indicate that TAR depends on DNA replication in human cells,Citation21,Citation22 suggesting that similar mechanisms operate in yeast and in higher eukaryotes. This view is supported by a recent report showing that depletion of Topoisomerase I (Top1) in mammalian cells induces replication fork stalling and DNA damage at highly-expressed genes.Citation38 Besides its well-characterized relaxation function, Top1 displays a kinase activity that is implicated in the regulation of splicing factors of the SR family such as ASF/SF2.Citation39 It has been shown that Top1 prevents interference between replication and transcription not only by relaxing topological constraints ahead of RNA and DNA polymerases but also by promoting ASF/SF2 function in order to prevent the formation of R-loops.Citation38 Interestingly, other mRNA-processing factors have been recently identified in a genome-wide siRNA screen for new factors preventing genomic instability.Citation40 As for Top1-deficient cells, spontaneous DNA damage was partially suppressed by RNase H in cells depleted for these factors, indicating that it is at least partly caused by R-loops.Citation38,Citation40 How R-loops interfere with DNA replication is currently unclear. RNA-DNA hybrids could directly impede the progression of the replisome. Alternatively, these structures could also promote the accumulation of DNA lesions on the non-coding strand, which would in turn interfere with DNA replication (). In both cases, it is tempting to speculate that these structures are able to interfere with DNA replication long after transcription has ceased. However, the stability of R-loop structures in the context of chromatin is currently unknown. Moreover, R-loop formation requires the presence of clusters of G on the non-transcribed strand, which may restrict their formation to specific regions of the genome.Citation41
Conclusion and Perspectives
Besides frontal collisions between replication and transcription machineries, a growing body of evidence indicates that cotranscriptional R-loops affect the integrity of eukaryotic genomes. Although these RNA-DNA hybrids play a positive role in developmentally-regulated processes such as class switch recombination at immunoglobulin genes,Citation2,Citation33 they also represent a major source of genomic instability.Citation15 Since patterns of gene expression and DNA replication are altered in oncogene-activated cells, it is tempting to speculate that transcription contributes to the replication stress observed in precancerous lesionsCitation42 and that cotranscriptional R-loops are involved in cancer development. Future experiments taking advantage of new ChIP-seq and RNA-seq technologies are required to test this hypothesis.
Abbreviations
| RNAP | = | RNA polymerase |
| TAR | = | transcription-associated recombination |
| DSB | = | DNA double-strand break |
| rDNA | = | ribosomal DNA |
| RFB | = | replication fork barrier |
| Top1 | = | topoisomerase I |
Figures and Tables
Figure 1 Models for interference between DNA replication and transcription. (A) The head-on collision model stipulates that a direct clash between the replisome and the RNA polymerase causes fork stalling, dissociation of the replisome and/or formation of recombinogenic reversed fork. This model implies that DNA replication and transcription occur simultaneously on the same DNA template. (B) In the cotranscriptional R-loops model, RNA-DNA hybrids formed during transcription interfere with replication fork progression and induce TAR. It is not clear whether fork arrest and recombination are caused by the DNA-RNA hybrid itself or by DNA lesions accumulating on the exposed ssDNA strand (diamonds).
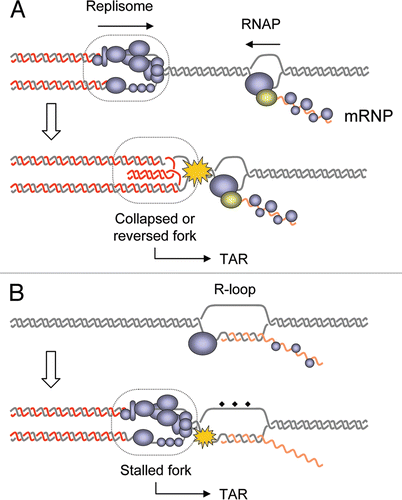
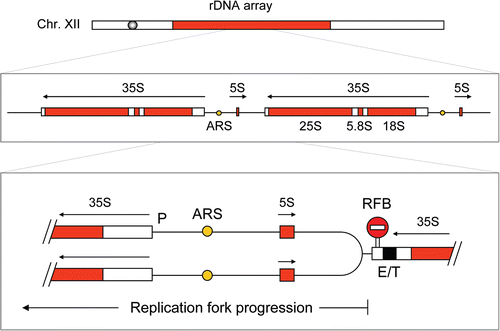
Figure 2 Schematic representation of the replication fork barrier at the S. cerevisiae rDNA array. This array is composed of ∼200 identical repeats (9.1 kb) containing a large 35S rRNA gene, a small 5S rRNA gene and a replication origin (ARS). Replication forks progressing opposite to the direction of 35S transcription are arrested at the replication fork barrier (RFB). P: Promoter of the 35S gene. E/T, enhancer/terminator.
Acknowledgements
We thank Armelle Lengronne and members of the PP laboratory for discussions and critical reading of the manuscript. A.M.P. was supported by an FRM post-doctoral fellowship. M.L.C. thanks Inserm for fellowship. Work in the P.P. laboratory is supported by FRM (Equipe FRM), ANR, INCa and the EMBO Young Investigator Programme.
References
- Mirkin EV, Mirkin SM. Replication Fork Stalling at Natural Impediments. Microbiol Mol Biol Rev 2007; 71:13 - 35
- Gottipati P, Helleday T. Transcription-associated recombination in eukaryotes: link between transcription, replication and recombination. Mutagenesis 2009; 24:203 - 210
- Liu B, Alberts BM. Head-on collision between a DNA replication apparatus and RNA polymerase transcription complex. Science 1995; 267:1131 - 1137
- Rothstein R, Michel B, Gangloff S. Replication fork pausing and recombination or “gimme a break.”. Genes Dev 2000; 14:1 - 10
- Vilette D, Uzest M, Ehrlich SD, Michel B. DNA transcription and repressor binding affect deletion formation in Escherichia coli plasmids. EMBO J 1992; 11:3629 - 3634
- Aguilera A. The connection between transcription and genomic instability. EMBO J 2001; 21:195 - 201
- Deshpande AM, Newlon CS. DNA replication fork pause sites dependent on transcription. Science 1996; 272:1030 - 1033
- Pomerantz RT, O'Donnell M. The replisome uses mRNA as a primer after colliding with RNA polymerase. Nature 2008; 456:762 - 766
- Pomerantz RT, O'Donnell M. Direct restart of a replication fork stalled by a head-on RNA polymerase. Science 2010; 327:590 - 592
- Rudolph CJ, Dhillon P, Moore T, Lloyd RG. Avoiding and resolving conflicts between DNA replication and transcription. DNA Repair 2007; 6:981 - 993
- Brewer BJ. When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cell 1988; 53:679 - 686
- Wang JD, Berkmen MB, Grossman AD. Genome-wide coorientation of replication and transcription reduces adverse effects on replication in Bacillus subtilis. Proc Natl Acad Sci USA 2007; 104:5608 - 5613
- Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell 1989; 56:619 - 630
- Nickoloff JA, Reynolds RJ. Transcription stimulates homologous recombination in mammalian cells. Mol Cell Biol 1990; 10:4837 - 4845
- Aguilera A, Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet 2008; 9:204 - 217
- Ivessa AS, Lenzmeier BA, Bessler JB, Goudsouzian LK, Schnakenberg SL, Zakian VA. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol Cell 2003; 12:1525 - 1536
- Prado F, Aguilera A. Impairment of replication fork progression mediates RNA polII transcription-associated recombination. EMBO J 2005; 24:1267 - 1276
- Takeuchi Y, Horiuchi T, Kobayashi T. Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev 2003; 17:1497 - 1506
- Azvolinsky A, Giresi PG, Lieb JD, Zakian VA. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol Cell 2009; 34:722 - 734
- Tourriere H, Pasero P. Maintenance of fork integrity at damaged DNA and natural pause sites. DNA Repair 2007; 6:900 - 913
- Gottipati P, Cassel TN, Savolainen L, Helleday T. Transcription-associated recombination is dependent on replication in mammalian cells. Mol Cell Biol 2008; 28:154 - 164
- Savolainen L, Helleday T. Transcription-associated recombination is independent of XRCC2 and mechanistically separate from homology-directed DNA double-strand break repair. Nucl Acids Res 2009; 37:405 - 412
- Kobayashi T, Horiuchi T. A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells 1996; 1:465 - 474
- Pasero P, Bensimon A, Schwob E. Single-molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus. Genes Dev 2002; 16:2479 - 2484
- Gerber JK, Gogel E, Berger C, Wallisch M, Muller F, Grummt I, et al. Termination of mammalian rDNA replication: polar arrest of replication fork movement by transcription termination factor TTF-I. Cell 1997; 90:559 - 567
- Huvet M, Nicolay S, Touchon M, Audit B, d'Aubenton-Carafa Y, Arneodo A, et al. Human gene organization driven by the coordination of replication and transcription. Genome Res 2007; 17:1278 - 1285
- Wei X, Samarabandu J, Devdhar RS, Siegel AJ, Acharya R, Berezney R. Segregation of transcription and replication sites into higher order domains. Science 1998; 281:1502 - 1505
- Vieira KF, Levings PP, Hill MA, Crusselle VJ, Kang S-HL, Engel JD, et al. Recruitment of transcription complexes to the {beta}-globin gene locus in vivo and in vitro. J Biol Chem 2004; 279:50350 - 50357
- Cook PR. The organization of replication and transcription. Science 1999; 284:1790 - 1795
- Kitamura E, Blow JJ, Tanaka TU. Live-cell imaging reveals replication of individual replicons in eukaryotic replication factories. Cell 2006; 125:1297 - 1308
- Drolet M. Growth inhibition mediated by excess negative supercoiling: the interplay between transcription elongation, R-loop formation and DNA topology. Mol Microbiol 2006; 59:723 - 730
- Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 2005; 122:365 - 378
- Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol 2003; 4:442 - 451
- Huertas P, Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell 2003; 12:711 - 721
- Li X, Manley JL. Cotranscriptional processes and their influence on genome stability. Genes Dev 2006; 20:1838 - 1847
- Gomez-Gonzalez B, Felipe-Abrio I, Aguilera A. The S-phase checkpoint is required to respond to R-loops accumulated in THO mutants. Mol Cell Biol 2009; 29:5203 - 5213
- Huertas P, Garcia-Rubio ML, Wellinger RE, Luna R, Aguilera A. A hpr1 point mutation that impairs transcription and mRNP biogenesis without increasing recombination. Mol Cell Biol 2006;
- Tuduri S, Crabbe L, Conti C, Tourriere H, Holtgreve-Grez H, Jauch A, et al. Topoisomerase 1 suppresses replication stress and genomic instability by preventing interference between replication and transcription. Nat Cell Biol 2009; 11:1315 - 1324
- Rossi F, Labourier E, Forne T, Divita G, Derancourt J, Riou JF, et al. Specific phosphorylation of SR proteins by mammalian DNA topoisomerase I. Nature 1996; 381:80 - 82
- Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell 2009; 35:228 - 239
- Roy D, Yu K, Lieber MR. Mechanism of R-loop formation at immunoglobulin class switch sequences. Mol Cell Biol 2008; 28:50 - 60
- Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science 2008; 319:1352 - 1355