Abstract
There are three stages of transcription: initiation, elongation and termination, and traditionally there has been a clear distinction between the stages. The specificity factor sigma is completely released from bacterial RNA polymerase after initiation, and then recycled for another round of transcription. Elongation factors then associate with the polymerase followed by termination factors (where necessary). These factors dissociate prior to initiation of a new round of transcription. However, there is growing evidence suggesting that sigma factors can be retained in the elongation complex. The structure of bacterial RNAP in complex with an essential elongation factor NusA has recently been published, which suggested rather than competing for the major σ binding site, NusA binds to a discrete region on RNAP. A model was proposed to help explain the way in which both factors could be associated with RNAP during the transition from transcription initiation to elongation.
The first step in gene expression, transcription, is one of the most highly regulated processes in the cell. In all organisms, transcription is carried out by a DNA-dependent RNA polymerase (RNAP). In eukaryotic cells, there are three distinct RNAPs: RNAP I transcribes rRNA precursors, RNAP II is responsible for the synthesis of mRNA and certain small nuclear RNAs and the synthesis of 5s rRNA and tRNA is performed by RNAP III.Citation1 In bacteria, there is only one RNAP responsible for the transcription of all classes of RNA. The evolutionarily conserved ∼400 kDa bacterial RNAP core enzyme (subunit composition α2ββ'ω) is capable of catalyzing RNA synthesis on its own. However, to initiate transcription RNAP core must be associated with a σ factor forming a holoenzyme, which is competent for specific binding to the promoter regions on DNA.Citation2 A typical promoter generally comprises two hexanucleotide consensus motifs: the −10 and −35 elements (nucleotide sequence: TTGACA and TATAAT, respectively) which are separated by a ∼17 base pair (bp) spacer.Citation3 Most bacteria have one primary housekeeping σ factor (σ70 in Escherichia coli and σA in Bacillus subtilis) and one or a number of alternative σ factors for transcribing specific classes of genes.Citation4 σ70-family members are both sequentially and structurally conserved among all bacteria and have four regions of homology designated 1–4, which can be further subdivided into evolutionarily conserved regions.Citation5 A DNA-binding domain of σ region 2, σ2.4, is responsible for sequence-specific interactions with the non-template strand of the −10 promoter element and σ factor region 4.2 (σ4.2) binds to the −35 promoter element.Citation6 The major RNAP-σ interface is formed between the region 2.2 of σ (σ2.2) and the solvent exposed clamp-helix (CH) region of the RNAP β' subunit ().Citation7,Citation8 Region 4 of the σ factor (σ4) also forms weak contacts with the flap domain (β flap) of the β subunit of RNAP, immediately adjacent to the tip of the RNA exit channel ().Citation9
There are three main sequential steps in the transcription cycle: promoter binding/initiation, RNA chain elongation and termination. Each step or intermediates within steps in the cycle represent a checkpoint for the regulation of gene expression by transcription factors.Citation10 To start transcription, RNAP scans the DNA () until a specific promoter region is recognized by the σ factor with σ2.4 binding to the −10 promoter element and σ4.2 binding to the −35 element. Subsequently ∼14 bp of DNA are melted around the AT-rich −10 box and then RNA synthesis is initiated at the +1 start site.Citation11 In most cases RNAP remains at the promoter region and initiates several rounds of abortive initiation, generating short transcripts 2–15 nucleotides (nt) in length.Citation12 Once about 12 nt of RNA have been synthesized, it is sufficiently long to fill the upstream RNA exit channel and destabilize the binding of σ region 4 to the β flap (). Disruption of the σ4-β flap interaction is the first step of σ dissociation from RNAPCitation9 and in turn triggers the release of the −35 promoter element. This allows RNAP to escape from the promoter and enter the elongation phase. It is widely accepted that after initiation factor σ completely dissociates from RNAP core, elongation factors become associated to regulate elongation and termination, followed by rebinding of σ to allow a new round of transcription initiation (the σ cycle).Citation13
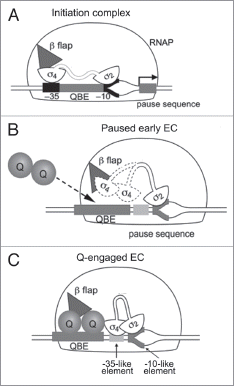
While the σ cycle is likely to be true most of the time, there is also some evidence that challenges the traditional view. σ factor has been shown to be present in a proportion of elongation complexes (ECs)Citation14,Citation15 and possibly plays functional roles in regulating transcription elongation (reviewed in ref. Citation13). The best-characterized system involves the formation of the bacteriophage λ Q protein antitermination complex required for the regulation of late gene transcription from the λ late promoter PR' (). There are two cis-acting sequences within the PR' regions required for the Q protein to engage the transcription machinery during the transition between initiation and elongation: a Q binding element (QBE) and a pause-inducing element (). As the nascent RNA transcribed from the PR' promoter emerges from the RNA exit channel at a length of ∼16 nucleotides, σ4 region is released from the β flap region and σ2 binds to the pause-inducing element (which resembles a promoter −10 element) and triggers pausing of the early EC (). The Q protein interacts with the QBE located between the promoter −10 and −35 elements, and engages this paused EC around the β flap region of RNAP. The Q protein also interacts with σ4 and stabilizes its binding to the −35-like pause-inducing element (). The σ factor is thus retained after RNAP has escaped the promoter and adopts a conformation in which both region 2 and 4 simultaneously bind the pause-inducing element (). This permits the Q protein to become a stable component of the EC, resulting in the formation of an antitermination complex, allowing RNAP to read through downstream transcription terminators and transcribe the phage λ late genes.Citation16,Citation17
The λ system exemplifies how σ factors are involved in the recruitment of elongation factors to RNAP. Another example we are focusing in this review is the interaction of the σ factor and elongation factor NusA with RNAP during the transition between initiation and elongation. NusA is highly conserved and essential in many organisms. It was first identified as the host factor involved in phage λ N-protein mediated antiterminationCitation18 and is also required in the antitermination complexes involved in host cell rRNA synthesis.Citation19 Conversely, NusA enhances RNAP pausing during transcription, particularly at pause sites, to ensure efficient intrinsic transcription termination.Citation20 NusA has also been shown to be involved in the “immune system” to suppress the toxic activity of foreign genes in E. coli host cells.Citation21 Structurally, NusA is a highly elongated molecule with the N-terminal domain (NTD) required for RNAP interaction joined by a flexible linker to the C-terminal S1/KH RNA binding domains.Citation22,Citation23
The traditional view of the transcription cycle involves the release of σ as RNAP clears the promoter, coincident with the binding of NusA and entry into the elongation phase of transcription. Results from several studies also showed that the binding of NusA and σ to RNAP was mutually exclusive, i.e., NusA cannot interact with RNAP when σ is bound and vice versa.Citation24 An α-helical region of the NusA NTD has also been observed to share structural homology with the major RNAP binding domain of σ factor (σ2), suggesting that the mutual exclusivity of binding resulted from direct competition of both factors for the same binding site on RNAP.Citation25 However, in vitro and in vivo results from other studies involving affinity isolation, single molecule and ChIP-chip analysis are inconsistent with the model due to the fact that both σ and NusA can be present simultaneously in transcription complexes under certain circumstances.Citation14,Citation15,Citation26,Citation27 Recently we proposed a model for RNAP in complex with NusA based on results from biochemical mapping and structural approaches.Citation28 Rather than competing for the major σ factor binding site (CH region of β'), NusA has been shown to bind principally to the β flap region via its NTD with the C-terminal domain (CTD) wrapped back across the β flap where it interacts with the emerging transcript.Citation28 The proposed NusA-RNAP complex model, which is consistent with other previous biochemical observations, has helped us better understand the various roles of NusA in the transcription cycle (also reviewed in ref. Citation29) and also enables the traditional σ cycle to be carefully re-examined.
In the initiation complex the σ2.4 and σ4.2 bind to the −10 and −35 promoter element respectively and in this conformation σ2.2 binds to the CH domain of the β' subunit and σ4 to the β flap (). The σ2-CH interaction has been shown to contribute to the majority of σ factor's affinity to RNAP, which could in turn stabilize the binding of σ4 to the β flap (Johnston EB and Lewis PJ; unpublished). Thus, NusA will not be able to compete with σ4 for its binding site on the β flap.Citation28 This is consistent with the fact that σ70 showed a much higher affinity for RNAP during initiation than NusA in a competition assay.Citation30 Once the promoter is cleared and σ4 is released from the β flap, NusA is able to become associated with RNAP at the β flap region via its NTD ( and D). Since the interactions between σ2.2 and the β' CH region of RNAP are not directly affected by σ4 displacement or NusA binding, a small fraction of σ could be retained. As a result, a transition complex from the initiation to elongation phase of transcription is formed containing σ bound to the CH region and NusA bound around the β flap, which has been previously reported ().Citation14 This is also consistent with recently published genome wide chromatin immunoprecipitation-microarray hybridization (ChIP-chip) data that shows that as σ slowly dissociates from the initiation complex after RNAP clears the promoter, NusA becomes associated, before the σ signal fully disappears.Citation27 Binding of NusA prevents σ4 from rebinding to the σ flap region, and also is likely to induce structural changes in RNAP resulting in the formation of a stable transcription EC, which in turn further reduces the affinity of σ interaction to RNAP, as observed previously.Citation30 In most cases, σ is gradually released from RNAP during elongation (). However, since the σ2.2 and β' CH interaction is not affected by NusA binding, under some circumstances σ could remain associated with (or rebind to) RNAP during elongation or through multiple rounds of transcription, as previously proposed.Citation13,Citation14 This is consistent with the ChIP-chip data that shows that a small proportion of the transcription complexes appear to retain σ throughout the cycle.Citation27
In summary, whilst the classic σ cycle, where σ completely dissociates from RNAP after initiation and rebinds for a new round of transcription, is likely to occur most of the time, there have been studies suggesting that this may not always be the case. σ factor has been shown to remain associated with the ECs and play regulatory roles under some circumstances. The recently published model of RNAP in complex with NusA has allowed us to refine the classic σ cycle and make an informed hypothesis on how the factors are interacting with RNAP during the transition from initiation to elongation.
Abbreviations
| bp | = | base pair |
| CH | = | clamp helix |
| CTD | = | C-terminal domain |
| ChIPchip | = | chromatin immunoprecipitation-microarray hybridization |
| EC | = | elongation complex |
| NTD | = | N-terminal domain |
| QBE | = | Q binding element |
| RNAP | = | RNA polymerase |
Figures and Tables
Figure 1 (A) Structural model of RNAP with the relative positions of the α, β, β' and ω subunits marked. The boxed areas indicate the clamp-helix (CH) region and β-flaps which are shown expanded below. σ region 2 binds the CH region, whilst σ region 4 and NusA NTD bind the β-flap. (B–E) Cartoon representation of the transcription machinery at different stages. (B) RNAP scans along the DNA until it encounters a promoter. (C) After a few rounds of abortive initiation, σ factor region 4 dissociates from the β flap, while region 2 remains bound to the CH region. (D) NusA binds to RNAP via its NTD while the rest of σ remains associated. (E) In most cases, σ is released from RNAP, resulting in a stable EC. See main text for detailed discussion.
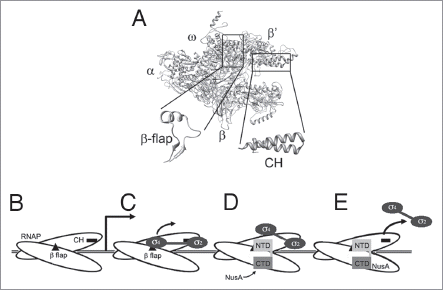
Figure 2 Formation of the phage Q protein antitermination complex at the PR' promoter. (A) The initiation complex with 2 bound to the −10 promoter element and 4 to the −35 promoter element. 4 is also shown interacting with the β flap (triangle) of RNAP. (B) The paused early EC at PR' with 2 bound to the pause-inducing −10-like element. 4 (dotted outline) is released from the β flap. (C) Q (shown as a dimer) binds to the QBE and become associated with the paused EC at the β flap region. Q also interacts with 4 and stabilizes its binding to the −35-like pausing element. Adapted from Nickels et al (ref. Citation16). See main text for detailed discussion.
Acknowledgements
Work in the lab of P.L. is supported by the ARC and NHMRC. X.Y. was supported by an Australian Postgraduate Award from the Australian Government.
References
- Archambault J, Friesen JD. Genetics of eukaryotic RNA polymerases I, II and III. Microbiol Rev 1993; 57:703 - 724
- Burgess RR, Anthony L. How sigma docks to RNA polymerase and what sigma does. Curr Opin Microbiol 2001; 4:126 - 131
- Estrem ST, Ross W, Gaal T, Chen ZW, Niu W, Ebright RH, et al. Bacterial promoter architecture: subsite structure of UP elements and interactions with the carboxy-terminal domain of the RNA polymerase alpha subunit. Genes Dev 1999; 13:2134 - 2147
- Ishihama A. Functional modulation of Escherichia coli RNA polymerase. Annu Rev Microbiol 2000; 54:499 - 518
- Lonetto M, Gribskov M, Gross CA. The σ70 family: sequence conservation and evolutionary relationships. J Bacteriol 1992; 174:3843 - 3849
- Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science 2002; 296:1285 - 1290
- Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, et al. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature 2002; 417:712 - 719
- Johnston EB, Lewis PJ, Griffith R. The interaction of Bacillus subtilis sigmaA with RNA polymerase. Protein Sci 2009; 18:2287 - 2297
- Nickels BE, Garrity SJ, Mekler V, Minakhin L, Severinov K, Ebright RH, et al. The interaction between sigma70 and the beta-flap of Escherichia coli RNA polymerase inhibits extension of nascent RNA during early elongation. Proc Natl Acad Sci USA 2005; 102:4488 - 4493
- Mooney RA, Artsimovitch I, Landick R. Information processing by RNA polymerase: recognition of regulatory signals during RNA chain elongation. J Bacteriol 1998; 180:3265 - 3275
- Murakami KS, Darst SA. Bacterial RNA polymerases: the wholo story. Curr Opin Struct Biol 2003; 13:31 - 39
- Goldman SR, Ebright RH, Nickels BE. Direct detection of abortive RNA transcripts in vivo. Science 2009; 324:927 - 928
- Mooney RA, Darst SA, Landick R. Sigma and RNA polymerase: an on-again, off-again relationship?. Mol Cell 2005; 20:335 - 345
- Bar-Nahum G, Nudler E. Isolation and characterization of sigma-retaining transcription elongation complexes from Escherichia coli. Cell 2001; 106:443 - 451
- Mukhopadhyay J, Kapanidis AN, Mekler V, Kortkhonjia E, Ebright YW, Ebright RH. Translocation of sigma with RNA polymerase during transcription: fluorescence resonance energy transfer assay for movement relative to DNA. Cell 2001; 106:453 - 463
- Nickels BE, Roberts CW, Roberts JW, Hochschild A. RNA-mediated destabilization of the sigma region 4/beta flap interaction facilitates engagement of RNA polymerase by the Q antiterminator. Mol Cell 2006; 24:457 - 468
- Deighan P, Diez CM, Leibman M, Hochschild A, Nickels BE. The bacteriophage lambda Q antiterminator protein contacts the beta-flap domain of RNA polymerase. Proc Natl Acad Sci USA 2008; 105:15305 - 15310
- Friedman DI, Baron LS. Genetic characterization of a bacterial locus involved in the activity of the N function of phage lambda. Virology 1974; 58:141 - 148
- Gourse RL, Gaal T, Bartlett MS, Appleman JA, Ross W. rRNA transcription and growth rate-dependent regulation of ribosome synthesis in Escherichia coli. Annu Rev Microbiol 1996; 50:645 - 677
- Schmidt MC, Chamberlin MJ. nusA protein of Escherichia coli is an efficient transcription termination factor for certain terminator sites. J Mol Biol 1987; 195:809 - 881
- Cardinale CJ, Washburn RS, Tadigotla VR, Brown LM, Gottesman ME, Nudler E. Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science 2008; 320:935 - 938
- Gopal B, Haire LF, Gamblin SJ, Dodson EJ, Lane AN, Papavinasasundaram KG, et al. Crystal structure of the transcription elongation/anti-termination factor NusA from Mycobacterium tuberculosis at 1.7 Å resolution. J Mol Biol 2001; 314:1087 - 1095
- Shin DH, Nguyen HH, Jancarik J, Yokota H, Kim R, Kim SH. Crystal structure of NusA from Thermotoga maritima and functional implication of the N-terminal domain. Biochemistry 2003; 42:13429 - 13437
- Gill SC, Weitzel SE, von Hippel PH. Escherichia coli sigma 70 and NusA proteins. I. Binding interactions with core RNA polymerase in solution and within the transcription complex. J Mol Biol 1991; 220:307 - 324
- Borukhov S, Lee J, Laptenko O. Bacterial transcription elongation factors: new insights into molecular mechanism of action. Mol Microbiol 2005; 55:1315
- Kapanidis AN, Margeat E, Laurence TA, Doose S, Ho SO, Mukhopadhyay J, et al. Retention of transcription initiation factor sigma70 in transcription elongation: single-molecule analysis. Mol Cell 2005; 20:347 - 356
- Mooney RA, Davis SE, Peters JM, Rowland JL, Ansari AZ, Landick R. Regulator trafficking on bacterial transcription units in vivo. Mol Cell 2009; 33:97 - 108
- Yang X, Molimau S, Doherty GP, Johnston EB, Marles-Wright J, Rothnagel R, et al. The structure of bacterial RNA polymerase in complex with the essential transcription elongation factor NusA. EMBO Rep 2009; 10:997 - 1002
- Yang X, Lewis PJ. The interaction between RNA polymerase and the elongation factor NusA. RNA Biol 2010; 7
- Gill SC, Weitzel SE, von Hippel PH. Escherichia coli sigma 70 and NusA proteins. I. Binding interactions with core RNA polymerase in solution and within the transcription complex. J Mol Biol 1991; 220:307 - 324