Abstract
Long non-coding RNAs (lncRNAs) have been shown to epigenetically regulate certain genes in human cells. Here we report evidence for the involvement of an antisense lncRNA in the transcriptional regulation of the pluripotency-associated factor Oct4. When an lncRNA antisense to Oct4-pseudogene 5 was suppressed, transcription of Oct4 and Oct4 pseudogenes 4 and 5 was observed to increase. This increase correlated with a loss of silent state epigenetic marks and the histone methyltransferase Ezh2 at the Oct4 promoter. We observed this lncRNA to interact with nucleolin and PURA, a 35-kD single-stranded DNA and RNA binding protein, and found that these proteins may act to negatively regulate this antisense transcript.
Introduction
Stem cells hold great potential as a research and therapeutic tool, but advancements in this technology have lagged due to ethical concerns regarding their isolation. Various research groups have recently described methods for reprogramming human and mouse somatic cells to induced pluripotent stem (iPS) cells.Citation1–Citation4 In human cells, reprogramming strategies generally involve introduction of some combination of pluripotency-associated factors, notably Oct4, Sox2, Lin28 and Nanog;Citation2 or Oct-3/4, Sox2, c-Myc and Klf4.Citation3,Citation4 Expression of these pluripotency-associated factors is important to maintaining the pluripotent state and the epigenetic silencing of these genes is an important step in differentiation.Citation5,Citation6 In particular, the methylation state of lysine 9 on histone 3 (H3K9) at the Oct4 promoter, as mediated by G9a and Jhmd2a, is an important factor in differentiation and reprogramming.Citation7–Citation9 Besides contributing to pluripotency in stem cells, Oct4, also known as POU5F1 and Oct3/4, has been implicated in oncogenesis. Such a link is appreciated due to the observation that cancer and stem cells share properties of self-renewal and propagation.Citation10
Genome-wide studies indicate that much more of the human genome is transcribed than previously realized, with many of these transcripts being long non-coding RNAs (lncRNAs).Citation11,Citation12 Many of these lncRNAs run antisense relative to protein-coding regions.Citation13,Citation14 Recent evidence from our group and others indicates that certain antisense lncRNAs function to regulate gene transcription by directing epigenetic remodeling complexes to particular loci.Citation15–Citation17 In these studies, knockdown of antisense transcripts by RNA interference (RNAi) led to activation of sense transcription. Antisense lncRNAs have also been shown to function in trans to direct epigenetic modifications to distal loci. In a study of lncRNAs associated with HOX loci in human cells, Rinn et al. found that an antisense RNA associated with the HOXC locus repressed transcription of the distal HOXD locus.Citation18 This antisense transcript was shown to associate with the Polycomb Repressive Complex 2 (PRC2) and the observed silencing correlated with an increase in H3K27me3 at the HOXD locus.
In mouse cells, lncRNAs have recently been identified which regulate Oct4 and Nanog, perhaps pointing to a role for non-coding transcripts in regulating pluripotency.Citation19 In human cells, Oct4 is expressed as two transcript variants which, through alternative splicing, differ in their 5′ untranslated regions (UTRs), 5′ coding regions and translational start codons.Citation20 In addition to these two transcript variants of Oct4, six related pseudogenes (Oct4-pg1-6) have also been identified in various human cell lines and cancers, although little is known regarding their function.Citation21 These pseudogenes were identified in cDNA samples, the reverse transcription of which was primed using an oligo(dT) primer. As the epigenetic regulation of Oct4 is important in many biologically relevant processes, we chose to investigate the role that these Oct4-related lncRNAs might play in the transcriptional regulation of Oct4. A detailed understanding of the epigenetic regulation of Oct4 is crucial if applications of iPS cells are to one day become therapeutically relevant. In addition, knowledge of RNA-based networks involved in regulating pluripotency may prove useful with regards to future methods of cellular reprogramming and the development of cancer-targeted therapeutics.
Results
Our initial characterization of Oct4-related transcripts employed several primer sets ( and Table S1). Oct4 primer set 2 specifically recognized the Oct4 promoter. Oct4 primer set 3 specifically recognized Oct 4 transcript variant 2. Oct4 primer set 1 recognized both transcript variants of Oct4, as well as Oct4 pseudogene 5 (Oct4-pg5). In addition, while the forward primer of set 1 also recognized Oct4-pg1 and 3 and the reverse primer also recognized Oct4-pg-4, these overlaps were not shared between primers, so PCR and qPCR analysis was not expected to be confounded by Oct4-pg1, 3 or 4. Previously published data show that of polyadenylated Oct4-related transcripts, only Oc4-pg5 is expressed in MCF-7 cells.Citation21 As such, sense Oct4-related transcripts measured using primer set 1 were understood to be Oct4-pg5. This previous screen did not, however, determine the presence or absence of non-polyadenylated lncRNAs that might span Oct4 or any of its pseudogenes.
To characterize the sense and antisense transcripts associated with Oct4, polymerase chain reaction (PCR) was used to analyze cDNA libraries generated by strand-specific reverse transcription PCR (RT-PCR). In strand-specific RT-PCR, reverse transcription was primed with either a gene-specific forward or reverse primer alone, thereby generating cDNA of specifically the antisense or sense strand of the targeted region respectively. PCR was then performed using forward and reverse primers, with amplicons representing sense or antisense transcripts which overlap that region. As published work has established that the only polyadenylated Oct4-related RNA expressed in MCF-7 cells is Oct4-pg5, these experiments were performed with poly(A)-depleted RNA.Citation21
Using each of our primer sets individually, we were able to detect non-polyadenylated RNAs antisense to Oct4 and/or Oct4-pg5 (primer set 1), overlapping the Oct4 promoter (primer set 2) and antisense to Oct4 transcript variant 2 (primer set 3) (). In each case, poly(A) depletion was confirmed by noting a lack of amplification of cDNA samples primed with oligo(dT). These data indicate that in MCF-7 cells, in addition to the previously described sense Oct4-pg5 transcript, there are also antisense transcripts overlapping the coding region of Oct4-pg5 and the promoter and coding regions of Oct4 (asOct4-pg5 and asOct4 respectively).
Strand-specific RT-PCR was also performed using primers specific for an intronic region of Oct4. Analysis with this primer set showed the presence of an antisense RNA overlapping this intronic region, but no sense transcript (Fig. S1). In addition to showing that the non-coding RNA in question overlaps intronic regions of Oct4, this result confirms the ability to selectively amplify target strands by the described method of strand-specific RT-PCR.
We hypothesized that the observed lncRNAs antisense to Oct4 and Oct4-pg5 may function to regulate Oct4-related sense transcripts. To test this hypothesis, we generated three siRNAs (siO18, siO78 and siO98; Table S1) targeted to asOct4 and asOct4-pg5 and assessed their effect on Oct4-related transcripts by quantitative PCR (qPCR) using primer set 1. In addition to targeting these asRNAs, the siRNAs screened could also target a putative lncRNA antisense to Oct4-pg4, although we were unable to detect an antisense RNA associated with Oct4-pg4 in MCF-7 cells (Fig. S1).
Transfection with siO18 and siO98 led to a suppression of asOct4 and/or asOct4-pg5 and a concurrent increase in Oct4 and/or Oct4-pg5 sense RNA levels (). This observed discordant regulation was similar to previous observations, where targeting antisense lncRNAs also resulted in increases in gene-specific sense/mRNA expression.Citation15–Citation17 These data suggest that Oct4-related transcripts may be in part controlled by an antisense lncRNAs associated with Oct4 and/or Oct4-pg5.
To further investigate the role that lncRNAs antisense to Oct4 and Oct4-pg5 might play in regulating Oct4-related sense RNA expression, we cloned siO18 and a scrambled control as short-hairpin RNAs (shO18 and shS18 respectively). Antisense Oct4 and/or Oct4-pg5 levels were shown to be decreased 48 h after transfection with shO18, and 72 h post-transfection we observed an increase in Oct4-related sense RNA levels ( and C). We also analyzed these samples using primer set 3, specific for Oct4 transcript variant 2. This analysis showed no decrease in the antisense transcript overlapping this region, but an increase in Oct4 mRNA (). These data suggest that asOct4-pg5 and not asOct4, is likely the primary effector molecule in this regulatory mechanism.
To more specifically assess the effect of shO18 treatment on Oct4-related transcripts, we next performed nuclear run-on. Analysis with primer set 1, specific for Oct4 and Oct4-pg5, showed an increase in Oct4 and/or Oct4-pg5 sense RNA transcription (). We next analyzed nuclear run-on samples using primers specific for Oct4 transcript variant 2 and Oct4 pseudogene 4 and we observed a significant increase in transcription of both these RNA species ( and C). These results indicate that the lncRNA asOct4-pg5 may function to regulate multiple Oct4-related transcripts. Values obtained from qPCR analysis of nuclear run-on samples were standardized first to actively transcribed β-actin and then to control samples (shS18).
In order to more fully characterize the mechanism by which asOct4-pg5 affects Oct4-related transcription, we used RNAi to knockdown various genes that have been implicated in epigenetic and RNA-mediated transcriptional regulation. We found that knockdown of the histone methyl transferases Ezh2 and G9a caused an increase in Oct4 mRNA, while knockdown of Ago1, HDAC1 and DNMT3a had no significant effect (). Values obtained from qPCR analysis were normalized first to β-actin and then to control samples (si854). Knockdown of these genes was confirmed by qPCR (Fig. S2A). While these data indicate that Ezh2 and G9a negatively regulate Oct4, they alone do not directly link this activity to asOct4-pg5.
To investigate the role that asOct4-pg5 might play in directing histone methyl transferases and chromatin modifications, we next assessed the effect of knockdown of asOct4-pg5 on chromatin structure at the Oct4 promoter. Chromatin immunoprecipitation (ChIP) analysis of the Oct4 promoter 72 h post-transfection with shO18 showed decreased levels of H3K9me2, H3K27me3 and Ezh2 (). These results indicate that asOct4-pg5 may function to recruit Ezh2 and possibly G9a to the promoter of Oct4, which in turn leads to an increase in silent state histone modifications and transcriptional silencing of Oct4. These data are strikingly similar to observations made when the respective antisense lncRNAs associated with p15, p21 and HOX loci were studied.Citation15,Citation16,Citation18
As little is known regarding mechanisms of RNA-mediated transcriptional regulation, we next sought to identify novel protein factors involved in this process by mass spectrometry. To do so we incubated nuclear extracts of MCF-7 cells with a biotin-tagged oligonucleotide designed to bind to asOct4 and/or asOct4-pg5 and pulled down the lncRNA and associated proteins with streptavidin beads (). Additional samples were incubated with a biotin-tagged irrelevant control oligonucleotide (miRN367) in order to monitor specificity of pull-down. The presence of the antisense RNA in question was validated by directional RT-PCR () and associated proteins were isolated and analyzed by mass spectrometry (). A full list of proteins identified in biotin-tagged control and O18 samples can be found in Table S2. Of the several proteins identified, we chose to further analyze purine-rich element binding factors A and B (PURA and PURB) and nucleolin (NCL). PURA, PURB and NCL are DNA and RNA binding proteins whose roles in gene regulation are widespread and in many ways ill-defined.Citation22–Citation25 As these proteins could interact with asOct4-pg5 or the targeted loci, they were considered of particular interest to our study.
We found that knockdown of PURA and NCL (Fig. S2B) had a negative effect on Oct4 mRNA levels (). Presented results were normalized first to β-actin and second to control samples (shS18). We also found that knockdown of PURA and NCL had no significant effect on levels of asOct4 and asOct4-pg5 (). These results suggested that PURA and NCL may function to inhibit the function of asOct4-pg5. To address this hypothesis, we analyzed changes at the Oct4 promoter upon shRNA-mediated knockdown of PURA and NCL. We found by ChIP analysis that knockdown of PURA resulted in an increase of H3K27me3 (). As asOct4-pg5 was previously shown to direct H3K27me3 to the Oct4 promoter (), this result may indicate that PURA inhibits this action through its interaction with asOct4-pg5, possibly by sequestering this asRNA away from targeted loci.
Discussion
While the mechanism of antisense lncRNA-mediated transcriptional regulation discussed here is similar to that of earlier described antisense lncRNAs, these findings are unique in that the antisense lncRNA in question lacks a poly A tail and originates from a pseudogene. Knockdown of asOct4-pg5 resulted in an increase in Oct4-pg5, as well as Oct4 and Oct4-pg4. This result may indicate that asOct4-pg5 has multiple targets or that multiple lncRNAs function together in some fashion to create a widespread effect on Oct4-related transcription.
Also presented here is evidence for novel functions of proteins which may regulate antisense lncRNAs. One protein found by mass spectrometry to associate with asOct4 and/or asOct4-pg5 is PURA. We observed that shRNA-mediated knockdown of PURA led to a decrease in Oct4 transcript levels and an enrichment of H3K27me3 at the Oct4 promoter. PURA is a multifunctional DNA- and RNA-binding protein which has been shown to regulate transcription, translation, cell cycle progression and cell proliferation.Citation22,Citation23 The data presented here suggest that PURA might also function to bind antisense RNAs and, possibly through interactions with additional proteins, sequester them away from their targets. PURB and NCL were also identified as asOct4 and/or asOct4-pg5 binding partners, although of these two, knockdown of only NCL had an effect on Oct4 transcripts. NCL is a ubiquitously expressed, multifunctional protein that plays a key role in ribosomal biogenesis.Citation24 NCL has also been observed to activate and repress gene transcription and to regulate RNA metabolism.Citation26 We found that while shRNA-mediated knockdown of NCL resulted in a decrease in Oct4 transcript levels, this effect did not correlate with changes in chromatin at the Oct4 promoter.
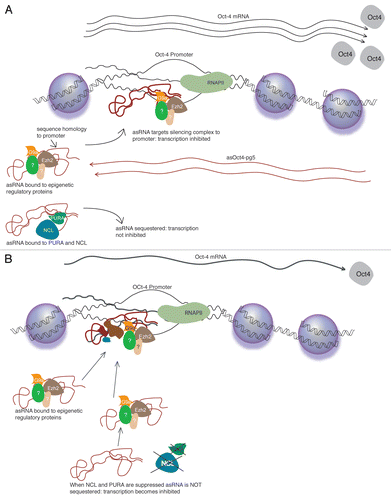
These results provide the basis for an expanded model of asRNA-mediated transcriptional regulation (). In this model, an asRNA associates with a complex of epigenetic regulatory proteins including Ezh2 and G9a. This complex is localized to a targeted promoter by the non-coding RNA. This complex then induces heterochromatization at the targeted promoter and subsequent silencing of transcription.Citation27 As this model suggests, we observed that RNAi-mediated knockdown of asOct4-pg5, Ezh2 or G9a led to increased transcription of Oct4. The proteins PURA and NCL function in this model to bind and sequester the asRNA. This sequestration renders the non-coding transcript inoperable, thereby blocking heterochromatization and transcriptional inhibition. In accordance with the model, we observed that RNAi-mediated knockdown of PURA and NCL resulted in decreased Oct4 mRNA levels.
The work presented here carries particular impact as Oct4 fulfills many important roles in the cell. Epigenetic regulation of Oct4 is important not only in reprogramming experiments, but also during differentiation of stem cells to progenitor cells and oncogenesis. These data provide insight into possible transcriptional regulators of Oct4 during such processes.
Materials and Methods
Cell culture and transfection.
Experiments were conducted in MCF-7 cells cultured at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and penicillin-streptomycin (Invitrogen™). Plasmid transfections were performed using Lipofectamine 2000 (Invitrogen™) at a concentration of 1 µg/1 × 106 cells and siRNA transfections were performed using Lipofectamine RNAiMAX (Invitrogen™) at a concentration of 100 nM.
Poly(A) depletion and PCR.
MCF-7 RNA was isolated (RNeasy, Qaigen™), DNase treated (TURBO DNase, Ambion™), depleted of polyadenylated transcripts (mRNA Catcher, Invitrogen™) and converted to cDNA (Reverse Transcrpition Core Kit, Eurogentec™) using indicated primers. cDNA was amplified by PCR (KAPA2G Fast HotStart, Kapa Biosystems) using indicated primers and analyzed on a 6% polyacrylamide gel. For a list of all primer sequences used in this work please see Table S1.
siRNA design and synthesis.
We designed and synthesized (Silencer siRNA Construction Kit, Ambion™) three siRNAs targeted to antisense RNAs associated with Oct4 and Oct4-pg5, and an irrelevant control siRNA targeted to CCR5 (si854). We also employed siRNAs targeted to Ago1, DNMT3a, Ezh2 and G9a, which, along with the control si854, were synthesized by Inegrated DNA Technologies™. Sequences of siRNAs used can be found in Table S1. Several of these siRNAs have been used and validated in previous reports.Citation28,Citation29
shRNA plasmid construction.
A short-hairpin RNA targeted to antisense RNAs associated with Oct4 and Oct4-pg5 and a scrambled control of this sequence were synthesized (Integrated DNA Technologies™) and cloned into the BLOCK-iT™ H1 RNAi Entry Vector (Invitrogen™) per manufacturer's guidlines. shRNA contructs targeting PURA (TRCN0000014534), PURB (TRCN0000155904) and NCL (TRCN0000062284) were obtained as a gift from Carol Kreider at Sigma-Aldrich™. Sequences of shRNAs used can be found in Table S1.
Quantitative PCR (qPCR).
RNA was extracted (RNeasy, Qaigen™), DNase treated (TURBO DNase, Ambion™), reverse transcribed using non-specific or indicated primer (Reverse Transcrpition Core Kit, Eurogentec™) and analyzed by qPCR using indicated primers (Kapa Sybr Fast, Kapa Biosystems™). Results from qPCR of cDNA generated in a strand-specific manner were normalized to a standard curve and then expressed as fractions of control values. Data from qPCR of cDNA generated non-specifically were normalized first to internal β-actin levels and then expressed as fractions of control values.
Chromatin immunoprecipitation (ChIP).
ChIP assay was performed as previously described.Citation30 DNA was immunopreciptated using an anti-H3K27me3 (Cell Signaling # 9756S), anti-H3K9me2 (Cell Signaling # 9753S), H3K4me2 (Upstate 07-030), anti-Ago1 (Santa Cruz Biotechnology # sc-32657), anti-Ezh2 (Upstate # 07-689) or anti-G9a (Cell Signaling #3306S) antibody and pulled down by Dynabeads Protein A (Invitrogen™). DNA was then recovered by phenol/chloroform extraction and analyzed by qPCR using indicated primers (Kapa Sybr Fast, Kapa Biosystems™).
Nuclear run-on.
Nuclear run-on was performed as previously described,Citation31 although analysis of run-on samples was slightly modified to use biotin-tagged transcripts.Citation28,Citation29 Biotinylated RNA was pulled down using Streptavidin-coupled Dynabeads (Invitrogen™). cDNA copies of pulled down RNA were generated (Reverse Transcription Core Kit, Eurogentec™) and then analyzed by qPCR using indicated primers (Kapa Sybr Fast, Kapa Biosystems™). Presented data were standardized first to beta-actin and subsequently to control (shS18) values.
Biotin pull-down.
MCF-7 cells were washed and lysed as described in previous ChIP assay. Lysed nuclei were incubated with a biotin labeled O18 or an irrelevant control (miRN367) oligonucleotide at a concentration of 500 µM for 30 minutes at 42°C (sequences of biotin-tagged oligonucleotides found in Table S1). Biotin-bound nucleic acids were pulled down using Streptavidin-coupled Dynabeads (Invitrogen™) and protein complexes were eluted by addition of 100 µl of 2× elution buffer (10 mM Tris-HCl (pH 6.0), 1 mM EDTA and 2.0 M NaCl) and incubation at 65°C for 10 min. Eluted proteins were analyzed by mass spectrometry (TSRI Center for Mass Spectrometry).
Abbreviations
| RNA | = | ribonucleic acid |
| iPS | = | induced pluripotent stem |
| Sox2 | = | SRY (sex-determining region Y)-box 2 |
| Klf4 | = | krüppel-like factor 4 |
| c-Myc | = | myelocytomatosis viral oncogene homolog |
| Lin28 | = | lin-28 homolog |
| Oct4 | = | octamer-binding transcription factor 4 |
| Oct4-pg | = | Oct4 pseudogene |
| POU5F1 | = | POU class 5 homeobox 1 |
| PURA | = | purine rich element binding protein A |
| PURB | = | purine rich element binding protein B |
| NCL | = | nucleolin |
| RNAi | = | RNA interference |
| mRNA | = | messenger RNA |
| lncRNA | = | long non-coding RNA |
| cDNA | = | complimentary DNA |
| asRNA | = | antisense RNA |
| miRNA | = | micro RNA |
| siRNA | = | small interfering RNA |
| H3K27me3 | = | trimethylated lysine 27 on histone 3 |
| H3K9me2 | = | dimethylated lysine 9 on histone 3 |
| Ago1 | = | argonaute 1 |
| HDAC1 | = | histone deacetylase 1 |
| DNMT3a | = | DNA methyltransferase 3a |
| Ezh2 | = | enhancer of zeste homolog 2 |
| G9a | = | euchromatic histone lysine N-methyltransferase 2 |
| HOX | = | homeobox gene |
| PCR | = | polymerase chain reaction |
| RT-PCR | = | reverse transcriptase PCR |
| qPCR | = | quantitative PCR |
Figures and Tables
Figure 1 (A) Schematic representation of transcripts, primer sites, siRNA target sites and other genetic elements associated with Oct4 and Oct4-pg5. (B) PCR of MCF-7 RNA depleted of polyadenylated transcripts and converted to cDNA in a strand-specific manner using indicated primers. PCR products were analyzed on 6% polyacrylamide gel. Non-polyadenylated antisense transcripts were detected overlapping the coding region of Oct4-pg5 and the coding and promoter regions of Oct4.
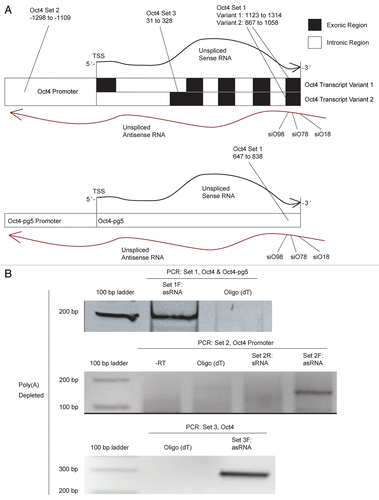
Figure 2 (A) Screen of three siRNAs targeted to lncRNAs antisense to Oct4 and Oct4-pg5. Compared to cells treated with the control siRNA, samples transfected with siO18 and siO98 demonstrated a decrease in antisense and an increase in sense transcripts associated with Oct4 and/or Oct4-pg5. (B) Forty-eight hours post-transfection with shO18, relative to samples treated with the scrambled control shS18, antisense transcripts associated with Oct4 and/or Oct4-pg5 were decreased, while sense transcripts were not significantly changed. (C) Seventy-two hours post transfection with shO18, antisense transcripts associated with Oct4 and/or Oct4-pg5 were decreased, while sense transcript levels were significantly increased. (D) Seventy-two hours post-transfection with shO18, antisense transcripts associated with Oct4 were unchanged, while sense transcript levels were significantly increased. (B–D) Averages are shown from cultures treated in triplicate, error bars represent standard errors of means and p values from two-sided t-tests are indicated.
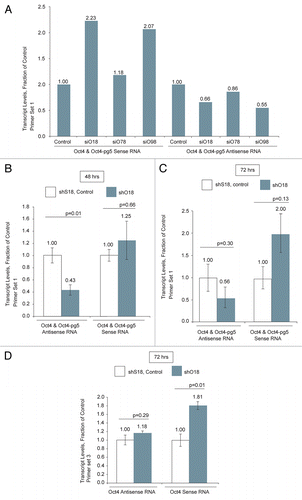
Figure 3 (A–C) Compared to cells treated with the scrambled control, samples transfected with shO18 demonstrated a significant increase in transcription of Oct4-pg5, Oct4 and Oct4-pg4. (D) Transfection of MCF-7 cells with siRNAs targeted to Ezh2 and G9a resulted in an increase in Oct4 sense transcript levels. (E) Treatment of MCF-7 cells with shO18 resulted in a loss of the silent state epigenetic marks H3K9me2 and H3K27me3 and the histone methyltransferase Ezh2 at the Oct4 promoter. (A–D) Values obtained from qPCR were standardized to β-actin levels. (A–E) Averages are shown from cultures treated in triplicate, error bars represent standard errors of means and p values from two-sided t-tests are indicated. Samples were analyzed 72 h post-transfection.
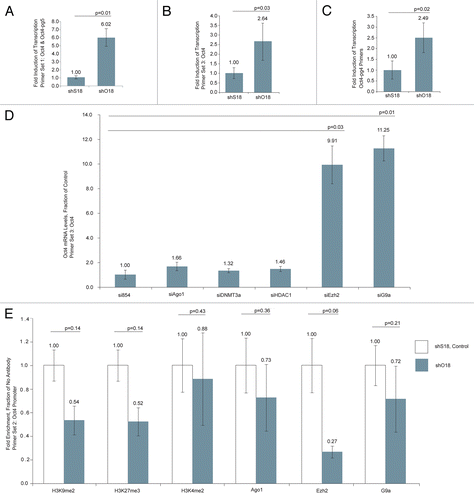
Figure 4 (A) Schematic of biotin-mediated pulldown of target asRNA. (B) RNA samples isolated from nuclei incubated with biotin-tagged O18 or control (miRN367) oligonucleotides were reverse transcribed with Oct4 Set 1 Fwd, amplified by PCR using Oct4 set 1 and analyzed on a 6% polyacrylamide gel. (C) Isolated proteins were analyzed by mass spectrometry and the indicated proteins, unique to samples pulled-down with biotin-tagged O18, were identified.
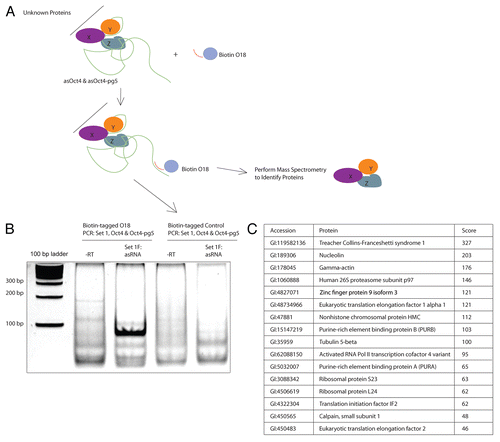
Figure 5 (A) Transfection of MCF-7 cells with shRNAs targeted to PURA and NCL resulted in a decrease in Oct4 sense transcript levels. Results have been normalized to internal β-actin levels. (B) Treatment of MCF-7 cells with a shRNA targeted to PURA resulted in an enrichment of the silent state epigenetic mark H3K27me3 at the Oct4 promoter. (C) Transfection of MCF-7 cells with shRNAs targeted to indicated genes resulted in no change in asOct4 or asOct4-pg5 levels. (A–C) Averages are shown from cultures treated in triplicate, error bars represent standard errors of means and p values from two-sided t-tests are indicated. Samples were analyzed 72 h post-transfection.
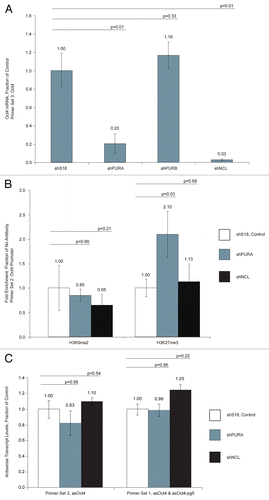
Figure 6 (A) The non-coding transcript asOct4-pg5 associates with Ezh2, G9a and possibly additional regulatory proteins. This complex localizes to the Oct4 promoter where it induces heterochromatization and inhibition of transcription. PURA and NCL may also bind asOct4-pg5, but function to inhibit its activity by sequestration. (B) When PURA and NCL are suppressed by RNAi, asOct4-pg5 is able to target Ezh2 and G9a to the Oct4 promoter, resulting in reduced Oct4 transcription.
Additional material
Download Zip (527.4 KB)Acknowledgements
This project is funded by NIH R01 HL083473 and NIH R01 AI084406 to K.V.M. We thank Carol Kreider, of Sigma-Aldrich, for providing shRNA constructs to suppress PURA, PURB and NCL.
References
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126:663 - 676
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007; 318:1917 - 1920
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature 2008; 451:141 - 146
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007; 131:861 - 872
- Adewumi O, Aflatoonian B, Ahrlund-Richter L, Amit M, Andrews PW, Beighton G, et al. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol 2007; 25:803 - 816
- Collas P, Noer A, Timoskainen S. Programming the genome in embryonic and somatic stem cells. J Cell Mol Med 2007; 11:602 - 620
- Feldman N, Gerson A, Fang J, Li E, Zhang Y, Shinkai Y, et al. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol 2006; 8:188 - 194
- Ma DK, Chiang CH, Ponnusamy K, Ming GL, Song H. G9a and Jhdm2a regulate embryonic stem cell fusion-induced reprogramming of adult neural stem cells. Stem Cells 2008; 26:2131 - 2141
- Epsztejn-Litman S, Feldman N, Abu-Remaileh M, Shufaro Y, Gerson A, Ueda J, et al. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol 2008; 15:1176 - 1183
- de Jong J, Looijenga LH. Stem cell marker OCT3/4 in tumor biology and germ cell tumor diagnostics: history and future. Crit Rev Oncog 2006; 12:171 - 203
- Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet 2006; 15:17 - 29
- Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science 2008; 319:1787 - 1789
- Conley AB, Miller WJ, Jordan IK. Human cis natural antisense transcripts initiated by transposable elements. Trends Genet 2008; 24:53 - 56
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, et al. Antisense transcription in the mammalian transcriptome. Science 2005; 309:1564 - 1566
- Morris KV, Santoso S, Turner AM, Pastori C, Hawkins PG. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet 2008; 4:1000258
- Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature 2008; 451:202 - 206
- Schwartz JC, Younger ST, Nguyen NB, Hardy DB, Monia BP, Corey DR, et al. Antisense transcripts are targets for activating small RNAs. Nat Struct Mol Biol 2008; 15:842 - 848
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007; 129:1311 - 1323
- Sheik Mohamed J, Gaughwin PM, Lim B, Robson P, Lipovich L. Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA 2010; 16:324 - 337
- Takeda J, Seino S, Bell GI. Human Oct3 gene family: cDNA sequences, alternative splicing, gene organization, chromosomal location and expression at low levels in adult tissues. Nucleic Acids Res 1992; 20:4613 - 4620
- Suo G, Han J, Wang X, Zhang J, Zhao Y, Dai J. Oct4 pseudogenes are transcribed in cancers. Biochem Biophys Res Commun 2005; 337:1047 - 1051
- Gallia GL, Johnson EM, Khalili K. Puralpha: a multifunctional single-stranded DNA- and RNA-binding protein. Nucleic Acids Res 2000; 28:3197 - 3205
- White MK, Johnson EM, Khalili K. Multiple roles for Puralpha in cellular and viral regulation. Cell Cycle 2009; 8:1 - 7
- Srivastava M, Pollard HB. Molecular dissection of nucleolin's role in growth and cell proliferation: new insights. FASEB J 1999; 13:1911 - 1922
- Wortman MJ, Johnson EM, Bergemann AD. Mechanism of DNA binding and localized strand separation by Pur alpha and comparison with Pur family member, Pur beta. Biochim Biophys Acta 2005; 1743:64 - 78
- Hanakahi LA, Dempsey LA, Li MJ, Maizels N. Nucleolin is one component of the B cell-specific transcription factor and switch region binding protein, LR1. Proc Natl Acad Sci USA 1997; 94:3605 - 3610
- Morris KV. Long antisense non-coding RNAs function to direct epigenetic complexes that regulate transcription in human cells. Epigenetics 2009; 4:296 - 301
- Hawkins PG, Santoso S, Adams C, Anest V, Morris KV. Promoter targeted small RNAs induce long-term transcriptional gene silencing in human cells. Nucleic Acids Res 2009; 37:2984 - 2995
- Turner AM, De La Cruz J, Morris KV. Mobilization-competent lentiviral vector-mediated sustained transcriptional modulation of HIV-1 expression. Mol Ther 2009; 17:360 - 368
- Han J, Kim D, Morris KV. Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proc Natl Acad Sci USA 2007; 104:12422 - 12427
- Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science 2004; 305:1289 - 1292