Abstract
Unlike prokaryotes, in which transcription and translation are coupled, eukaryotes physically separate transcription in the nucleus from mRNA translation and degradation in the cytoplasm. However, recent evidence have revealed that the full picture is more complex, and that the nuclear transcription machinery plays specific roles in regulating the cytoplasmic fate of mRNA.
The crosstalk between transcription, translation and degradation of mRNA can be deduced intuitively for prokaryotes, since transcription and translation occur in the same time and space. Eukaryotes, on the other hand, segregate their chromosomes from the rest of the cell and the barrier of the nuclear membrane divides transcription from protein expression. It is now well established that transcription and nuclear mRNA processing are coupled, as mRNA maturation (capping, splicing and polyadenylation) and mRNA export are initiated on nascent transcripts.Citation1 Even if a process like spliceosome assembly can be initiated during transcription, evidence suggest that some genes are spliced cotranscriptionally, while for other genes splicing occurs post-transcriptionally.Citation2,Citation3 Moreover, these events depend on interactions between the transcription machinery and RNA processing factors, in which the transcription elongation complex acts as a platform to recruit these factors.Citation4 It is known that several RNA-binding proteins which specify the cytoplasmic fate of transcripts (i.e., translational control, mRNA decay or mRNA localization) shuttle between the cytoplasm and the nucleus, and their presence in the nucleus is essential for their function.Citation5 For instance, the exon-junction complex (E°C), which is deposited on mRNA after splicing, follows transcripts to the cytoplasm, where it enhances mRNA translation, targets transcripts to nonsense-mediated decay (NMD) and participates in mRNA localization.Citation5 However, it is still unclear to what extent these factors are recruited during transcription of their target mRNA, and how this occurs. Here, we review current evidence showing how the RNA polymerase II transcription machinery directly affects mRNA translation, localization or degradation in the cytoplasm by promoting the cotranscriptional recruitment of RNA-binding proteins involved in these processes. This evidence comes mostly from the budding yeast, in which these pathways have been well studied. However, since several of the factors involved are highly conserved throughout evolution, these mechanisms are likely relevant to higher eukaryotes as well.
Rpb4/7 Couples Transcription and Cytoplasmic mRNA Degradation
RNA polymerase II is constituted of 12 subunits in eukaryotes, including ten core subunits, which encompass the catalytic site, and the Rbp4-Rbp7 subcomplex (Rpb4/7), which forms a stalk-like protrusion on the RNA polymerase II core, in close proximity to the RNA exit channel.Citation6 Rpb7 is known to bind the nascent RNA, although with little specificity.Citation6 In yeast, deletion of RPB7 is lethal, while Rpb4 is essential for viability only at high temperature.Citation7 Unlike RNA polymerase II core subunits, Rpb4 and Rbp7 are essential only for transcription initiation in vitro, and are dispensable for transcription elongation.Citation8 Genome wide chromatin immunoprecipitation in yeast revealed that Rpb4/7 occupancy is identical to Rpb3, suggesting that this heterodimer is associated with RNA polymerase II throughout the transcription cycle.Citation9 Like most small subunits of RNA polymerase II in yeasts (S. cerevisiae and S. pombe), Rpb4 is in excess compared to the large subunits Rpb1, Rpb2 and Rpb3.Citation10,Citation11 Moreover, Rpb4 and Rpb7 (and Rpb3 in humans cellsCitation12) are the only subunits that actively shuttle between the nucleus and the cytoplasm,Citation13 suggesting other roles for Rpb4/7 beside transcription.
Indeed, Rpb4/7 has been found to affect the cytoplasmic degradation of mRNA in yeast. In the cytoplasm, Rpb4 enhances the degradation of specific transcripts, called protein biosynthetic factors mRNA (PBF mRNA) as they encode for ribosomal proteins or translational factors.Citation14 The turnover of PBF mRNA is extremely sensitive to environmental stimuli, and Rpb4 regulates their deadenylation and degradation rate in different conditions. Rpb4 interacts physically and genetically with components of the major 5′ to 3′ mRNA degradation pathway in yeast, like Pat1 and Lsm2, and accumulates in cytoplasmic bodies called P bodies, which are sites of mRNA storage and degradation.Citation15 A role for Rpb7 in cytoplasmic mRNA decay has also been demonstrated.Citation16 The generation of thermosensitive mutants of Rpb7 in yeast revealed that some mutants displayed a defect in mRNA decay, and not in transcription, at the non-permissive temperature. These Rpb7 mutants compromised the degradation of both PBF and non-PBF transcripts, suggesting a more general role for Rpb7 in mRNA decay, compared to Rbp4. Unlike Rpb4, Rpb7 acts via both 5′ to 3′ and 3′ to 5′ mRNA degradation pathways, as it interacts physically and genetically with both pathways. While these results suggested a function for both Rpb4 and Rpb7 in cytoplasmic mRNA decay, it was still unclear if this function depends on the association of Rpb4/7 with the core subunits of RNA polymerase II.
Evidence emerged on the implication of the transcription machinery in Rpb4/7-dependent cytoplasmic mRNA decay.Citation17 Specific thermosensitive mutants of RNA polymerase II (Rpb6Q100R and Rpb1C67S, C70S), which disrupt interactions with the Rpb4/7 heterodimer, display defect in mRNA degradation similar to those observed previously for Rpb4 and Rpb7 mutants. This phenotype is specific for these rpb6 and rpb1 alleles, since other alleles of RPB1 do not display a defective mRNA decay. Overexpression of Rpb4/7 partially rescues the defect in mRNA decay observed in the Rpb6Q100R mutant, suggesting that the interaction between Rpb4/7 and RNA polymerase II is important for the role of Rpb4/7 in cytoplasmic mRNA degradation. These results strongly suggest that the cotranscriptional recruitment of Rpb4/7 by RNA polymerase II is necessary for its role in cytoplasmic mRNA decay, arguing against the possibility that the Rpb4/7 heterodimer can be recruited to mRNA independently of the RNA polymerase. While the role of Rpb4/7 in mRNA decay has been observed mostly with transcripts that are part of the PBF family, little is known about its role in the general degradation of mRNA. It would be interesting to investigate if this mechanism applies to all mRNA.
Mechanistic Coupling Between Transcription and Translation
Besides its role in cytoplasmic mRNA decay, a recent publication revealed another function for the Rpb4/7 complex in promoting efficient translation initiation.Citation18 Indeed, yeast Rpb4/7 was found to physically interact with Nip1 and Hcr1, two components of the translation initiation factor 3 (eIF3), which serves as a platform for the recruitment of the small ribosomal subunit during translation initiation.Citation19 Deletion of RPB4 and a conditional mutant allele of RPB7 (rpb7–26) show reduced formation of polysomes and hypersensitivity to translation inhibitors. Importantly, the rpb7–26 allele does not display any major defects in transcription or mRNA decay, suggesting that the effects observed on translation are not indirect. The role of Rpb4/7 in translation initiation was particularly evaluated during the stimulation of translation following exit from glucose starvation. During glucose starvation, mRNA is excluded from polysomes and stored in P bodies. When glucose is added back to yeast culture, the mRNA move from P bodies to the polysomes and resume translation.Citation20 This process was significantly reduced in the rpb7–26 mutant and RPB4 deletion strains, demonstrating an implication of Rpb4/7 in translation initiation.
An important point in this study was that the role of Rpb4/7 in translation initiation depends on its interaction with the core RNA polymerase II. Indeed, mutants defective in interaction of RNA pol II with Rpb4/7 (Rpb6Q100R and Rpb1C67S, C70S) also exhibit abnormal pattern of translation. Furthermore, nuclear import of Rpb4 is essential to this new function, since a NLS-mutated Rpb4 displayed abnormal polysome profile. From these results, a model can be proposed in which RNA polymerase II regulates translation initiation by stimulating the binding of Rpb4/7 on nascent mRNA in the nucleus, before they reach the ribosomes and initiate translation in the cytoplasm.
Based on this study and on previous ones revealing a role in mRNA degradation, the Rpb4/7 heterodimer has been coined as an “mRNA coordinator” as this complex interacts with mRNA at each step of their life cycle—from transcription, translation to mRNA decay (). However, these results raise questions concerning the function of this coordination between the transcription machinery, translation and mRNA decay. A key observation is that the role of Rpb4/7 in these processes is particularly important during environmental stress conditions, like heat stress and glucose starvation.Citation18 Indeed, while Rpb7 is an essential gene, Rpb4 is essential for transcription during temperature stress.Citation21 Furthermore, Rpb4 is known to mediate nuclear export of mRNA during stress conditions and most Rpb4 molecules accumulate in the cytoplasm during stress.Citation22 Altogether, these data suggest a role for Rpb4/7 in integrating cellular response to environmental stress by acting on mRNA, from their synthesis in the nucleus to their translation and decay in the cytoplasm.
Spt4-Spt5 Link Transcription to Cytoplasmic mRNA Localization
Beside translation and mRNA decay, recent evidence point toward a novel role for the transcription machinery in promoting cytoplasmic mRNA localization. mRNA localization is a mechanism which restricts spatial and temporal mRNA translation in a cell. This process is essential for the establishment of cellular polarity, synaptic plasticity, asymmetric cell division and cell fate determination.Citation23 Evidence from several organisms suggests that the mRNA localization process is initiated on nascent mRNAs. For instance, the RNA-binding proteins ZBP1 and ZBP2, which are involved in β-actin mRNA localization at the leading edge in fibroblasts, are recruited on nascent β-actin mRNA.Citation24,Citation25 Localization of oskar mRNA at the posterior pole of the Drosophila embryo requires members of the exon junction complex (EJC), like Y14-Mago and eIFIIIA.Citation26,Citation27 While the assembly of the EJC takes place concomitantly with splicing and can occur cotranscriptionally, at least in mammalian cells,Citation28 it is not clear yet if EJC assembly occurs cotranscriptionally in Drosophila. In both cases, it is still unclear if this recruitment is coupled to the transcription machinery, as it was shown for nuclear pre-mRNA processing pathways.
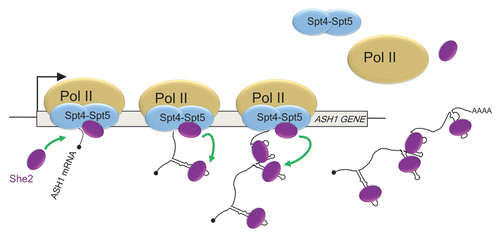
A link between transcription and mRNA localization has been more clearly established in the budding yeast, in which over 30 transcripts are actively transported and localized to the bud tip.Citation29 mRNA localization to the bud depends on the RNA-binding protein She2, which forms with She3 and Myo4 a complex called the “locasome” that carries their cargo along the actin filaments to the bud tip.Citation30 She2 recognizes specific RNA motifs, called localization elements or zipcodes, present within the coding sequence or 3′ untranslated region of localized transcripts.Citation31 While it was already known that She2 associates with mRNA in the nucleus,Citation32 recent evidence have made clear that She2 is actively imported in the nucleus via a non-classical nuclear localization signal (NLS), and that disrupting nuclear import of She2 (by mutating its NLS or retaining it in the cytoplasm) perturb mRNA localization at the bud and asymmetric protein synthesis.Citation33,Citation34
A role for the transcription machinery in the cotranscriptional recruitment of She2 to nascent bud-localized transcripts has been recently revealed.Citation35 Using chromatin immunoprecipitation (ChIP), She2 was found to be associated with genes coding for bud-localized mRNA, in a transcription-dependent manner. Treatment with RNase or a mutation in the RNA-binding domain of She2 reduces its association with these genes, suggesting that She2 binds the nascent transcripts. Interestingly, She2 interacts in vivo with the elongating form of RNA polymerase II, raising the possibility that She2 may be recruited to chromatin via the transcription machinery. Indeed, She2 directly interacts with the transcription factor Spt4–Spt5, also known as DSIF, which is part of the transcription elongation complex.Citation36 Spt4 and Spt5 form an heterodimeric complex which regulates the processivity of RNA pol II.Citation37 Beside its role in elongation, Spt5 is also implicated in mRNA capping, as it interacts and activates the cap guanylyltransferase,Citation38 and in mRNA splicing.Citation39 Mutations in either SPT4 or SPT5 reduce the cotranscriptional recruitment of She2 on a gene coding for a bud-localized mRNA and disrupt its localization at the bud. This leads to a model in which Spt4–Spt5 serves as a “launching pad,” which favors the cotranscriptional loading of She2 on nascent transcripts ().
It is well understood that the cotranscriptional recruitment of mRNA processing factors imposes a temporal order on the different steps of pre-mRNA maturation (from 5′ end capping to mRNA export) and mediates quality control of transcripts before nuclear export.Citation4 In comparison, it is not yet clear what the advantages for the cotranscriptional loading of RNA-binding proteins regulating cytoplasmic events, like translation, mRNA localization or decay, could be. One possibility is that interactions with the transcription machinery may ensure that RNA-binding factors specifying the cytoplasmic fate of an mRNA are loaded before mRNA export. Evidence from yeast, Drosophila and vertebrate show that, in the absence of their mRNA localization factors or translational repressors, localized mRNA are still exported from the nucleus.Citation26,Citation34,Citation40 This suggests the absence of a nuclear quality control that would retain or degrade improperly loaded transcripts before their export to the cytoplasm, which would cause their premature translation, degradation or mislocalization. The cotranscriptional recruitment of factors which regulate the cytoplasmic fate of mRNA may help avoid this problem by loading these factors prior transcription termination.
Another possibility is that cotranscriptional recruitment allows an integration of different regulatory mechanisms before nuclear export of an mRNA. Rpb4/7 is a good example, as it regulates both mRNA translation and decay. It is also known that the presence of She2 in the nucleus is essential for the recruitment of the translational repressor Puf6 on the bud-localized ASH1 mRNA in yeast.Citation34 Cotranscriptional recruitment of She2 on nascent mRNA may favor the subsequent loading of Puf6, and ensure that translational control of bud-localized transcripts is established before their export to the cytoplasm. It will be of interest to determine if other pathways mediating the cytoplasmic fate of mRNA are also coupled with transcription, and to what extent the transcription machinery primes mRNA for translational regulation, localization or decay in the cytoplasm.
Figures and Tables
Figure 1 Rpb4/7 is involved in transcription, translation and mRNA decay. Rpb4/7 associates with the core RNA polymerase II (pol II) subunits during elongation and is loaded cotranscriptionally on specific transcripts. In the cytoplasm, Rpb4/7 recruits eIF3 and promotes translation initiation. During stress, Rpb4/7 follows mRNA to P bodies and favors the return of transcripts to polysomes after stress. In addition, Rpb4/7 can promote the degradation of its target mRNA, via polyA tail shortening, followed by exonuclease degradation. As Rpb4/7 actively shuttles between the nucleus and cytoplasm, it can be re-imported to the nucleus.
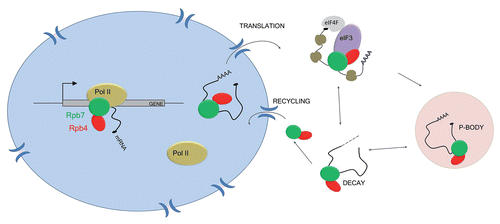
Figure 2 Spt4–Spt5 promote the cotranscriptional recruitment of She2 on the nascent ASH1 mRNA. Interaction between She2 and Spt4–Spt5 on elongating RNA polymerase II (pol II) favors the transfer of She2 on localization elements RNA as they emerge from the polymerase. The bud-localized ASH1 mRNA contains four localization elements, which fold as stem-loop structures.
Acknowledgements
We thank Emmanuelle Querido for critical reading of the manuscript. This work was supported by the grant MOP43855 from the Canadian Institutes for Health Research (CIHR) to P.C. P.C is a Senior Scholar from the Fonds de la Recherche en Santé du Québec (FRSQ).
References
- Komili S, Silver PA. Coupling and coordination in gene expression: a systems biology view. Nat Rev Genet 2008; 9:38 - 48
- Pandya-Jones A, Black DL. Co-transcriptional splicing of constitutive and alternative exons. RNA 2009; 15:1896 - 1908
- Tardiff DF, Lacadie SA, Rosbash M. A genome-wide analysis indicates that yeast pre-mRNA splicing is predominantly posttranscriptional. Mol Cell 2006; 24:917 - 929
- Perales R, Bentley D. Cotranscriptionality: The transcription elongation complex as a nexus for nuclear transactions. Mol Cell 2009; 36:178 - 191
- Giorgi C, Moore MJ. The nuclear nurture and cytoplasmic nature of localized mRNPs. Semin Cell Dev Biol 2007; 18:186 - 193
- Ujvari A, Luse DS. RNA emerging from the active site of RNA polymerase II interacts with the Rpb7 subunit. Nat Struct Mol Biol 2006; 13:49 - 54
- Woychik NA, Young RA. RNA polymerase II subunit RPB4 is essential for high- and low-temperature yeast cell growth. Mol Cell Biol 1989; 9:2854 - 2859
- Edwards AM, Kane CM, Young RA, Kornberg RD. Two dissociable subunits of yeast RNA polymerase II stimulate the initiation of transcription at a promoter in vitro. J Biol Chem 1991; 266:71 - 75
- Jasiak AJ, Hartmann H, Karakasili E, Kalocsay M, Flatley A, Kremmer E, et al. Genome-associated RNA polymerase II includes the dissociable Rpb4/7 subcomplex. J Biol Chem 2008; 283:26423 - 26427
- Rosenheck S, Choder M. Rpb4, a subunit of RNA polymerase II, enables the enzyme to transcribe at temperature extremes in vitro. J Bacteriol 1998; 180:6187 - 6192
- Kimura M, Sakurai H, Ishihama A. Intracellular contents and assembly states of all 12 subunits of the RNA polymerase II in the fission yeast Schizosaccharomyces pombe. Eur J Biochem 2001; 268:612 - 619
- Boulon S, Pradet-Balade B, Verheggen C, Molle D, Boireau S, Georgieva M, et al. HSP90 and its R2TP/prefoldin-like cochaperone are involved in the cytoplasmic assembly of RNA polymerase II. Mol Cell 39:912 - 924
- Selitrennik M, Duek L, Lotan R, Choder M. Nucleocytoplasmic shuttling of the Rpb4p and Rpb7p subunits of Saccharomyces cerevisiae RNA polymerase II by two pathways. Eukaryot Cell 2006; 5:2092 - 2103
- Lotan R, Bar-On VG, Harel-Sharvit L, Duek L, Melamed D, Choder M. The RNA polymerase II subunit Rpb4p mediates decay of a specific class of mRNAs. Gene Dev 2005; 19:3004 - 3016
- Franks TM, Lykke-Andersen J. The control of mRNA decapping and P-body formation. Mol Cell 2008; 32:605 - 615
- Lotan R, Goler-Baron V, Duek L, Haimovich G, Choder M. The Rpb7p subunit of yeast RNA polymerase II plays roles in the two major cytoplasmic mRNA decay mechanisms. J Cell Biol 2007; 178:1133 - 1143
- Goler-Baron V, Selitrennik M, Barkai O, Haimovich G, Lotan R, Choder M. Transcription in the nucleus and mRNA decay in the cytoplasm are coupled processes. Gene Dev 2008; 22:2022 - 2027
- Harel-Sharvit L, Eldad N, Haimovich G, Barkai O, Duek L, Choder M. RNA polymerase II subunits link transcription and mRNA decay to translation. Cell 2010; 143:552 - 563
- Hinnebusch AG. eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem Sci 2006; 31:553 - 562
- Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 2005; 310:486 - 489
- Choder M. Rpb4 and Rpb7: subunits of RNA polymerase II and beyond. Trends Biochem Sci 2004; 29:674 - 681
- Farago M, Nahari T, Hammel C, Cole CN, Choder M. Rpb4p, a subunit of RNA polymerase II, mediates mRNA export during stress. Mol Biol Cell 2003; 14:2744 - 2755
- Martin KC, Ephrussi A. mRNA localization: Gene expression in the spatial dimension. Cell 2009; 136:719 - 730
- Oleynikov Y, Singer RH. Real-time visualization of ZBP1 association with β-Actin mRNA during transcription and localization. Curr Biol 2003; 13:199 - 207
- Pan F, Huttelmaier S, Singer RH, Gu W. ZBP2 facilitates binding of ZBP1 to β-Actin mRNA during transcription. Mol Cell Biol 2007; 27:8340 - 8351
- Hachet O, Ephrussi A. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature 2004; 428:959 - 963
- Palacios IM, Gatfield D, St. Johnston D, Izaurralde E. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature 2004; 427:753 - 757
- Custodio N, Carvalho C, Condado I, Antoniou M, Blencowe BJ, Carmo-Fonseca M. In vivo recruitment of exon junction complex proteins to transcription sites in mammalian cell nuclei. RNA 2004; 10:622 - 633
- Shepard KA, Gerber AP, Jambhekar A, Takizawa PA, Brown PO, Herschlag D, et al. Widespread cytoplasmic mRNA transport in yeast: Identification of 22 bud-localized transcripts using DNA microarray analysis. Proc Natl Acad Sci USA 2003; 100:11429 - 11434
- Paquin N, Chartrand P. Local regulation of mRNA translation: new insights from the bud. Trends Cell Biol 2008; 18:105 - 111
- Olivier C, Poirier G, Gendron P, Boisgontier A, Major F, Chartrand P. Identification of a conserved RNA motif essential for She2p recognition and mRNA localization to the yeast bud. Mol Cell Biol 2005; 25:4752 - 4766
- Kruse C, Jaedicke A, Beaudouin J, Bohl F, Ferring D, Guttler T, et al. Ribonucleoprotein-dependent localization of the yeast class V myosin Myo4p. J Cell Biol 2002; 159:971 - 982
- Du TG, Jellbauer S, Müller M, Schmid M, Niessing D, Jansen RP. Nuclear transit of the RNA-binding protein She2 is required for translational control of localized ASH1 mRNA. EMBO Rep 2008; 9:781 - 787
- Shen Z, Paquin N, Forget A, Chartrand P. Nuclear shuttling of She2p couples ASH1 mRNA localization to its translational repression by recruiting Loc1p and Puf6p. Mol Biol Cell 2009; 20:2265 - 2275
- Shen Z, St-Denis A, Chartrand P. Cotranscriptional recruitment of She2p by RNA pol II elongation factor Spt4-Spt5/DSIF promotes mRNA localization to the yeast bud. Gene Dev 2010; 24:1914 - 1926
- Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Gene Dev 1998; 12:343 - 356
- Hartzog GA, Wada T, Handa H, Winston F. Evidence that Spt4, Spt5 and Spt6 control transcription elongation by RNA polymerase II in Saccharomy cescerevisiae. Gene Dev 1998; 12:357 - 369
- Wen Y, Shatkin AJ. Transcription elongation factor hSPT5 stimulates mRNA capping. Gene Dev 1999; 13:1774 - 1779
- Lindstrom DL, Squazzo SL, Muster N, Burckin TA, Wachter KC, Emigh CA, et al. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol Cell Biol 2003; 23:1368 - 1378
- Huttelmaier S, Zenklusen D, Lederer M, Lorenz M, Dictenberg J, Meng X, et al. Spatial regulation of beta-actin mRNA translation by Src-dependent phosphorylation of ZBP1. Nature 2005; 438:512 - 515