Abstract
In response to environmental and nutritional stimuli, a whole array of proteins remodel genome architecture, activate or transcribe genes, suppress genes, repair lesions and base-modifications, faithfully replicate and safely separate the parental and daughter genomes during cell division. Negotiating and resolving conflicts of genome trafficking is essential for genome stability.
Head-On Versus Co-Directional Replication Transcription Collisions
Genome trafficking drives cellular life. A multitude of proteins engage with various loci across the genome in response to a variety of environmental and nutritional stimuli. This continuous interplay of proteins with the DNA produces a dynamic, active genome with inevitable conflicts between different functions that must be safely negotiated and resolved to maintain genome stability. Hence, safe and accurate passage of the genetic information from the parental to the daughter cell is ensured. In bacteria, such conflicts are exacerbated by the need to respond quickly to stimuli. Under fast growth conditions, replication, transcription, translation, repair and recombination may all be functioning simultaneously increasing the potential for conflicts. One of the best studied and most fundamental conflict is that between DNA replication and transcription.Citation1–Citation4
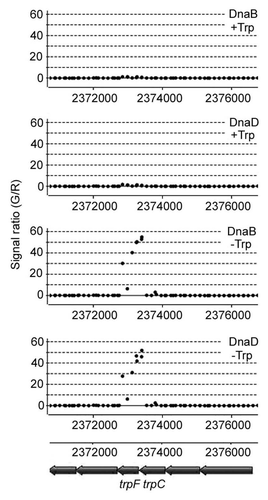
Both functions are carried out by complex molecular machines that translocate along the same DNA substrate and inevitable conflicts will arise when the two machines encounter each other along the DNA lattice. As a consequence of the polar 3′-5′ translocation of the RNAP (RNA polymerase) along the DNA substrate (lagging strand relative to replication) head-on traffic collisions with the replicative helicase moving in the opposite direction (5′-3′) along the same lagging strand are inevitable (). Head-on collisions are thought to be destructive for both the transcription and the replication molecular machines and this is one of the reasons why, in bacterial genomes, genes proximal to the replication origin (oriC) and highly expressed genes are co-directionally aligned with the clock- or anticlock-wise replisomes.Citation5 However, co-directional encounters also occur, particularly as replication is 10–20 fold faster than transcription.Citation2 When a fast replicative helicase, moving 5′-3′ along the lagging strand, catches up with a slower RNAP moving 3′–5′ along the opposite, anti-parallel, leading strand co-directional collisions are inevitable (). Co-directional encounters were, until recently, considered non-deleterious for the replication fork. However, Merrikh et al.Citation6 recently showed that, despite the fact that Bacillus subtilis ribosomal RNA (rRNA) genes are co-directionally aligned relative to replication, they are hot spots for conflicts that lead to the intervention of the replication restart machinery under fast growing conditions in vivo. Merrikh and co-workers observed transcription-dependent, DnaA-independent and PriA-dependent association of the essential replication restart proteins DnaD, DnaB and of the replicative helicase DnaC at rRNA loci in fast growing but not in slow growing cells. Using a similar ChIP-chip approach we found that in the B. subtilis strain 168, which is auxotrophic for tryptophan,Citation7 both DnaB and DnaD are associated with the trp operon in fast growing cultures when there is no tryptophan supplement in the media but not when the media are supplemented with tryptophan (40 mg/mL), (). It is now becoming evident that highly expressed chromosomal regions are potential hot spots for replication-transcription conflicts irrespective of relative directionality.
Ribosomal RNA Loci are Natural Impediments for Replication
In fast-growing bacteria, rRNA is highly expressed to sustain the high rates of protein synthesis. Multiple RNAPs are transcribing rRNA in tandem to sustain the increased cellular demand and spontaneous pausing events can lead to severe traffic jamsCitation8 that could potentially pose formidable obstacles to replication. rRNA genes are co-directionally aligned with replication in virtually all bacteria. Indeed, the first in vivo evidence that head-on replication collisions are problematic was provided in a seminal paper by French.Citation9 An IPTG-inducible ColE1 replication origin was inserted on either side of the Escherichia coli rrnB gene and electron microscopy analysis of DNA samples 4 and 6 min post-induction revealed that replication forks moving co-directionally to transcription from an upstream ColE1 origin towards rrnB were able to pass through the coding region, while forks moving from a downstream ColE1 origin head-on to transcription were delayed within the coding rrnB region. In Saccharomyces cerevisiae, rRNA genes were also shown to be natural impediments to replication.Citation10 Human cells have approximately 400 copies of rRNA genes in tandem repeats distributed over the short arms of several chromosomes.Citation11,Citation12 They are highly expressed with rRNA comprising as much as 70–80% of the total cellular RNA. Intriguingly, only a proportion (∼50%) of these repeats is transcribed in actively growing cells in interphase. Such selective transcription may be a necessary mechanism to resolve traffic conflicts between replication and transcription by allowing simultaneous progression of replication unimpeded through the silent regions.
The importance of avoiding collisions between replication and transcription in rRNA sites is highlighted by the evolution of ribosomal fork barriers (RFBs) found in many species including yeast, mice and humans.Citation2,Citation13 Proteins such as Fob1 in S. cerevisiae, Reb1 and Sap1 in Schizosaccharomyces pombe and TTF1 in mammals bind to sites peripheral to rRNA loci and form robust RFBs for replication forks. RFBs prevent the replication fork from entering the rRNA regions when they are transcriptionally active. Alternatively, unusual DNA structures also form formidable RFBs and serve the same purpose. Although the accumulation of RNAP in highly expressed rrn regions poses obstacles to replication, paradoxically large chromosomal inversions containing up to 4 rrn operons in E. coli do not significantly affect viability even when the main homologous recombination protein RecA is absent, suggesting that accessory proteins enable the replication fork to pass through such highly expressed regions.Citation14 However, chromosomal inversions of rrn genes render E. coli reliant on DinG whose function may be to remove R-loops that form within highly expressed rrn regions and thus block replication.Citation15
Negotiating Genome Trafficking
Clearing the pathway ahead of the replication fork is conceptually the simplest mechanism of resolving conflicts. The bacterial transcription elongation factors GreA and GreB clear the pathway by reactivating backtracked, stalled RNAPs. They interact with RNAP and coordinate a Mg2+ ion in the active site promoting hydrolysis of the trapped transcript.Citation16 The transcriptional terminator Rho also plays a role in clearing the path of RNAPs ahead of the replication fork. Inhibition of Rho-dependent transcription by bicyclomycin has been shown to induce double strand breaks, characteristic of replication fork arrest, whilst cells deleted for the Rho-cofactors NusA and NusG became hypersensitive to bicyclomycin and exhibited excessive chromosomal breaks.Citation17 The transcription-repair coupling factor Mfd performs a similar function by translocating along the DNA and displacing or nudging forward stalled RNAPs.Citation18,Citation19 Once a stalled RNAP is cleared out of the way, direct restart of the replication fork can resume DNA synthesis assuming that the stalled fork remains fully functional.Citation20 Despite the fact that Mfd plays a role in resolving replication-transcription traffic conflicts, cells lacking Mfd still exhibit normal growth. Recombination-mediated repair and other RNAP modulators may explain this lack of growth defects but cells lacking Rep and UvrD have severe growth problems in rich media irrespective of the fact that Mfd is present,Citation21 suggesting that Mfd cannot compensate for the role(s) of Rep and UvrD in resolving such conflicts. Only in slow growth conditions can this phenotype be suppressed. Rep and UvrD helicases exhibit 3′-5′ translocation polarity (opposite to the 5′-3′ polarity of the replicative ring helicase) and may thus act along the leading strand at replication forks to clear the way ahead. It remains to be established whether such accessory helicases are an integral part of the replication fork or specifically recruited to stalled replication forks as part of the direct recombination-free fork restart mechanism.
In vitro experiments with reconstituted replication forks using purified proteins suggested that co-directional collisions of replication-transcription result in displacement of the RNAP, leaving an intact replication fork that uses the RNA transcript as a primer to resume DNA synthesis.Citation22 Although such mechanism may indeed be true in vitro, the situation in vivo is drastically different, with multiple RNAPs in tandem under fast growth conditions transcribing the same gene and forming several mRNAs simultaneously. It is difficult to envisage how the replication fork will be able to displace a whole array of tandem RNAPs and remain intact to use multiple mRNAs as primers. It is also questionable whether the DNA-mRNA bubble remains intact in vivo when the RNAP is displaced. Indeed, forward displacement of RNAP by Mfd and Rho results in unwinding of the DNA-mRNA hybrid and release of the mRNA.Citation23
An alternative mechanism of resolving co-directional conflicts may involve a replication fork slowing down behind a co-directionally moving RNAP until transcription is completed before resuming rapid DNA synthesis. E. coli replication forks have a half life of 4–6 minCitation24,Citation25 and in some cases they may be stable even up to 1 hour.Citation20 Indeed, in simple cases when the RNAP is only temporarily halted the fork may stall and remain fully functional while waiting for transcription to be completed. The question still remains whether in fast growing conditions replication has the “luxury” of waiting around for transcription. A more stubborn challenge is posed by backtracked RNAP complexes, which are extremely stable, up to several hours (possibly days).Citation26 Waiting around for backtracked RNAPs to be cleared on their own accord will pose significant time delays and is not a viable option for replication. They will inevitably need to be swiftly cleared from the path of the replication fork. Preventing the formation of backtracked, stalled RNAPs will be beneficial for replication. Concomitant translation under fast growth conditions may have an important role to play in this respect. The rate of transcription in vivo is tightly controlled by the rate of translation. Higher rates of ribosomal protein synthesis accelerate the RNAP and prevent spontaneous backtracking.Citation27 Such translation-mediated acceleration of transcription could potentially facilitate the progression of the replication fork through sites of high activity in the case of co-directional encounters but could also potentiate the destructive effect between the replication fork and RNAP in head-on collisions. It is not, however, relevant to rRNA genes because they are not used for protein synthesis.
Replication Restart Mechanisms
The mechanism of replication restart is likely dictated by the nature of the conflict. Conflicts may come in the form of stationary or moving protein roadblocks, single or double strand breaks in the DNA or even DNA-mediated roadblocks in the form of stem-loops and other complex DNA secondary structures. Broadly speaking, replication restart mechanisms can be classified into recombination-free and recombination-mediated. Several studies have demonstrated direct interplay between replication and recombination, and have been reviewed by Rudolph et al.Citation1 A key goal that future studies should aim for is to precisely understand what happens to replication forks when they encounter variable conflicts. In some conflicts the replication fork can remain fully functional and simply restart DNA synthesis once the conflict is resolved but in many cases the replication fork may be deactivated by partial or total collapse and will need to be re-activated. Determining the composition(s) of stalled forks will provide a better understanding of whether certain partially collapsed forks can reactivate themselves or whether they need assistance by replication restart proteins. A key component of the replication fork, that may define the terms partial or total collapse, is the replicative ring helicase. It is conceivable that as long as the ring helicase remains tethered around the lagging strand, the stalled fork could simply be able to recruit any missing components without assistance. If, however, the helicase is displaced from the DNA, re-activation of the fork will require the intervention of the helicase-loading machinery to reload the helicase on the lagging strand.
Two precise restart mechanisms have been defined in E. coli. A PriA-mediated mechanism restarts replication in DNA substrates with a single stranded gap on the lagging strand, whilst a PriC-mediated mechanism operates in substrates with a single stranded gap on the leading strand.Citation28 PriA is a ubiquitous bacterial protein that recognizes and binds to the 3′-end of the nascent leading strand of the stalled forkCitation29 stabilizing it and preventing the unreplicated arm from being unwound. In the absence of a 3′-nascent end at the fork, PriA abortively unwinds the unreplicated arm causing fork instability. RecG prevents PriA from carrying out abortive unwinding.Citation30 Mutations in the priA gene causing deficiency of PriA-helicase were identified as suppressors of the recG phenotype suggesting that, in the absence of RecG, the PriA helicase activity produces deleterious effects, probably related to abortive unwinding of stalled replication forks. RecG regresses the fork to produce the characteristic “chicken-foot” structures, which are processed further by RuvABC, RecBCD and RecA to promote the remodelling of the fork and to introduce a 3′-end at the fork for PriA binding. The helicase activity of PriA is redundant in direct restart but required in PriC-mediated restart. The prominent role of PriA in resolving replication-transcription conflicts is highlighted by the increased association of replication restart proteins with the replicative helicase in highly expressed rRNA sites when PriA is depleted, and by the genetic interaction between priA mutants and RNAP mutants that affect RNAP translocation or stability.Citation6
Co-directional collisions of replication and transcription along the lagging strand are likely to result in DNA substrates with a single stranded gap on the lagging strand, previously occupied by the ring helicase and a 3′-end on the nascent leading strand near the branch onto which PriA will bind. Hence, the simplest, and possibly the fastest, restart mechanism to resolve co-directional conflicts will be PriA-mediated direct restart. It remains the case that head-on encounters are potentially more deleterious because the helicase and the RNAP collide directly on the lagging strand, whereas in the case of co-directional encounters the two motors could potentially bypass each other as they are moving on opposite strands. In such cases the leading strand polymerase could conceivably leave the fork to allow bypassing of the RNAP and could rejoin the fork further downstream. The resultant single stranded gap left on the leading strand could be filled in at a later stage once RNAP is out of the way. At sites of exceptionally high transcription activity such simple bypass mechanism may not be possible and fork collapse leading to the intervention of the restart mechanism could be the only option. More generally, the nature of the restart mechanism will very much depend on the nature of the conflict and the DNA substrate that is left behind. All cells have evolved important auxiliary replication restart proteins and an array of recombination-mediated mechanisms to safely negotiate a variety of conflicts in order to complete accurate genome duplication. Recombination-mediated restart mechanisms are inherently more precarious if not coordinated tightly, while direct restart mechanisms are safer in maintaining genomic stability.
Figures and Tables
Figure 1 Schematic representation of head-on and co-directional collisions between replication and transcription. In head-on collisions the replicative helicase and the RNAP translocate along the same strand (lagging strand) but in opposite directions. In co-directional collisions the helicase and the RNAP translocate in the same direction but along opposite strands, lagging and leading strands, respectively.
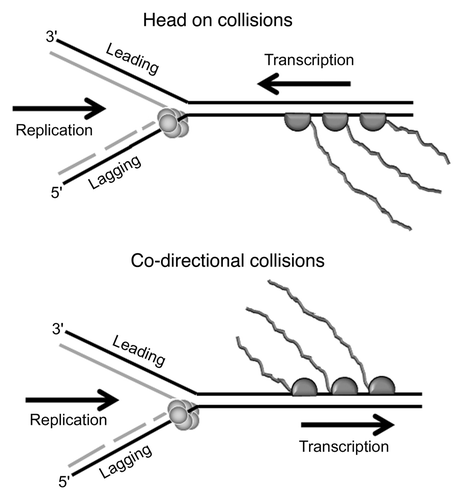
Figure 2 Association of the replication restart proteins with the trp locus in B. subtilis strain 168, grown in the presence and absence of tryptophan. Samples from cultures grown in the presence or absence of tryptophan (40 mg/mL) in rich media were analysed by ChIP-chip, as described by Merrikh et al.Citation6 Strong association of DnaD and DnaB with the trp locus was detected only when cells were grown in the absence of tryptophan, indicating that the transcriptional activation of the trp locus results in replication-transcription conflicts that lead to the intervention of the replication restart machinery.
Acknowledgments
Research in the P.S. laboratory is supported by a Biotechnology and Biological Sciences Research Council grant (BB/E006450/1) and a Wellcome Trust grant (091968/Z/10/Z).
References
- Rudolph CJ, Dhillon P, Moore T, Lloyd RG. Avoiding and resolving conflicts between DNA replication and transcription. DNA Repair 2007; 6:981 - 993; PMID: 17400034; http://dx.doi.org/10.1016/j.dnarep.2007.02.017
- Mirkin EV, Mirkin SM. Replication fork stalling at natural impediments. Microbiol Mol Biol Rev 2007; 71:13 - 35; PMID: 17347517; http://dx.doi.org/10.1128/MMBR.00030-06
- Aladjem MI. Replication in context: dynamic regulation of DNA replication patterns in metazoans. Nat Rev Cenet 2007; 8:588 - 600; PMID: 17621316; http://dx.doi.org/10.1038/nrg2143
- Pomerantz RT, O'Donnell M. What happens when replication and transcription complexes collide?. Cell Cycle 2010; 9:2535 - 2541; PMID: 20581460; http://dx.doi.org/10.4161/cc.9.13.12122
- Rocha EP. The replication-related organization of bacterial genomes. Microbiology 2004; 150:1609 - 1627; PMID: 15184548; http://dx.doi.org/10.1099/mic.0.26974-0
- Merrikh H, Machón C, Grainger WH, Grossman AD, Soultanas P. Co-directional replication-transcription conflicts lead to replication restart. Nature 2011; 470:554 - 557; PMID: 21350489; http://dx.doi.org/10.1038/nature09758
- Zeigler DR, Pràgai Z, Rodriguez S, Chevreux B, Muffler A, Albert T, et al. The origins of 168, W23 and other Bacillus subtilis legacy strains. J Bacteriol 2008; 190:6983 - 6995; PMID: 18723616; http://dx.doi.org/10.1128/JB.00722-08
- Klumpp S, Hwa T. Stochasticity and traffic jams in the transcription of ribosomal RNA: Intriguing role of termination and antitermination. Proc Natl Acad Sci USA 2008; 105:18159 - 18164; PMID: 19017803; http://dx.doi.org/10.1073/pnas.0806084105
- French S. Consequences of replication fork movement through transcription units in vivo. Science 1992; 258:1362 - 1365; PMID: 1455232; http://dx.doi.org/10.1126/science.1455232
- Azvolinski A, Giresi PG, Lieb JD, Zakian VA. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Molecular Cell 2009; 23:722 - 734; PMID: 19560424; http://dx.doi.org/10.1016/j.molcel.2009.05.022
- Birch JL, Zomerdijk JC. Structure and function of ribosomal RNA gene chromatin. Biochem Soc Trans 2008; 36:619 - 624; PMID: 18631128; http://dx.doi.org/10.1042/BST0360619
- Huang S, Rothblum LI, Chen D. Ribosomal chromatin organization. Biochem Cell Biol 2006; 84:444 - 449; PMID: 16936818; http://dx.doi.org/10.1139/O06-089
- Tsang E, Carr AM. Replication fork arrest, recombination and the maintenance of ribosomal DNA stability. DNA Repair 2008; 7:1613 - 1623; PMID: 18638573; http://dx.doi.org/10.1016/j.dnarep.2008.06.010
- Ensault E, Valens M, Espeli O, Boccard F. Chromosome structuring limits genome plasticity in Escherichia coli. PLoS Genetics 2007; 3:226; PMID: 18085828; http://dx.doi.org/10.1371/journal.pgen.0030226
- Boubakri H, de Septenville AL, Viguera E, Michel B. The helicases DinG, Rep and UvrD cooperate to promote replication across transcription units in vivo. EMBO J 2009; 29:145 - 157; PMID: 19851282; http://dx.doi.org/10.1038/emboj.2009.308
- Opalka N, Chlenov M, Chacon P, Rice WJ, Wriggers W, Darst SA. Structure and function of the transcription elongation factor GreB bound to bacterial RNA polymerase. Cell 2003; 114:335 - 345; PMID: 12914698; http://dx.doi.org/10.1016/S00928674(03)00600-7
- Washburn RS, Gottesman ME. Transcription termination maintains chromosome integrity. Proc Natl Acad Sci USA 2011; 108:792 - 797; PMID: 21183718; http://dx.doi.org/10.1073/pnas.1009564108
- Deaconescu AM, Chambers AL, Smith AJ, Nickels BE, Hochschild A, Savery NJ, Darst SA. Structural basis for bacterial transcription-coupled DNA repair. Cell 2006; 124:507 - 520; PMID: 16469698; http://dx.doi.org/10.1016/j.cell.2005.11.045
- Park JS, Marr MT, Roberts JW. E. coli transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell 2002; 109:757 - 767; PMID: 12086674; http://dx.doi.org/10.1016/S0092-8674(02)00769-9
- Pomerantz RT, O'Donnell M. Direct restart of a replication fork stalled by head-on RNA polymerase. Science 2010; 327:590 - 592; PMID: 20110508; http://dx.doi.org/10.1126/science.1179595
- Guy CP, Atkinson J, Gupta MK, Mahdi AA, Gwynn EJ, Rudolph CJ, et al. Rep provides a second motor at the replisome to promote duplication of protein-bound DNA. Molecular Cell 2009; 36:564 - 566; PMID: 19941825; http://dx.doi.org/10.1016/j.molcel.2009.11.009
- Pomerantz RT, O'Donnell M. The replisome uses mRNA as a primer after colliding with RNA polymerase. Nature 2008; 456:762 - 766; PMID: 19020502; http://dx.doi.org/10.1038/nature07527
- Park JS, Roberts JW. Role of DNA bubble rewinding in enzymatic transcription termination. Proc Natl Acad Sci USA 2006; 103:4870 - 4875; PMID: 16551743; http://dx.doi.org/10.1073/pnas.0600145103
- Marians KJ, Hiasa H, Kim DR, McHenry CS. Role of the core DNA polymerase III subunits at the replication fork. A is the only subunit required for processive replication. J Biol Chem 1998; 273:2452 - 2457; PMID: 9442096; http://dx.doi.org/10.1074/jbc.273.4.2452
- McGlynn P, Guy CP. Replication forks blocked by protein-DNA complexes have limited stability in vitro. J Mol Biol 2008; 381:249 - 255; PMID: 18602646; http://dx.doi.org/10.1016/j.jmb.2008.05.053
- Uptain SM, Kane CM, Chamberlin MJ. Basic mechanisms of transcript elongation and its regulation. Ann Rev Biochem 1997; 66:117 - 172; PMID: 9242904; http://dx.doi.org/10.1146/annurev.biochem.66.117
- Proshkin S, Rahmouni AR, Mironov A, Nudler E. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science 2010; 328:504 - 508; PMID: 20413502; http://dx.doi.org/10.1126/science.1184939
- Heller RC, Marians KJ. The disposition of nascent strands at stalled replication forks dictates the pathway of replisome loading during restart. Molecular Cell 2005; 17:733 - 743; PMID: 15749022; http://dx.doi.org/10.1016/j.molcel.2005.01.019
- Sasaki K, Ose T, Okamoto N, Maenaka K, Tanaka T, Masai H, et al. Structural basis of the 3′-end recognition of a leading strand in stalled replication forks by PriA. EMBO J 2007; 26:2584 - 2593; PMID: 17464287; http://dx.doi.org/10.1038/sj.emboj.7601697
- Rudolph CJ, Upton AL, Briggs GS, Lloyd RG. Is RecG a general guardian of the bacterial genome?. DNA Repair 2010; 9:210 - 223; PMID: 20093100; http://dx.doi.org/10.1016/j.dnarep.2009.12.014