Abstract
The persistence of a reservoir of transcriptionally competent but latent virus in the presence of antiviral regimens presents the main impediment to a curative therapy against HIV. Therefore it is critical to understand the molecular mechanisms, which lead to the establishment and maintenance of HIV latency, and which contribute to the reversal of this process and mediate HIV transcriptional activation in response to T cell activation signals. Here I discuss features of the nucleosomal landscape of the HIV promoter or 5'LTR in controlling HIV transcription. I emphasize on the emerging understanding of the role of the ATP dependent SWI/SNF chromatin remodelling complexes in modulating the chromatin architecture at the HIV LTR and how this leads to a tight regulation of LTR transcription.
When the Human Immunodeficiency Virus type-l (HIV-1) infects its host cell, it integrates into the genome as a chromatin template. Through unclear mechanisms, a very small percentage of integrated HIV establishes a transcriptionally silent infection in resting memory CD4 T cells, the main reservoir of latent provirus.Citation1 This latent reservoir is masked from combination Antiretroviral Therapy (c-ART), which is extremely effective against the active virus. However, because it is replication competent, latent HIV retains the capacity to become transcriptionally activated, leading to increased viral load upon treatment interruption.Citation1 Thus, the persistence of this pool of transcriptionally competent but silenced HIV reservoir presents the main impediment to a curative therapy. It is therefore crucial to elucidate the underlying molecular mechanisms that control the establishment and maintenance of HIV latency, and which contribute to the activation of the silenced integrated HIV promoter. Here, I discuss the nucleosomal architecture of the HIV promoter (or 5′LTR), with an emphasis on the role played by the SWI/SNF chromatin remodeling complexes in modulating the nucleosomal landscape and thereby transcription at the HIV LTR.
Chromatin Organization And Transcription At The HIV LTR
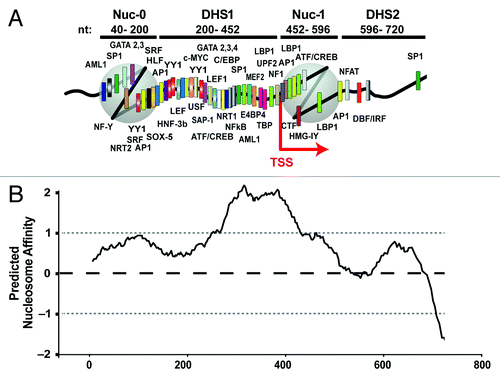
Transcription of the HIV genome is controlled by the HIV promoter or 5′LTR. Irrespective of the position of HIV-1 integration within the host genome, the 5′LTR is organized into precisely positioned nucleosomes in its basal repressed transcriptional state (). The repressed HIV-1 proviral 5′ LTR is organized into two positioned nucleosomes, nuc-0 and nuc-1, that are connected by an intervening enhancer region, which is hypersensitive to digestion by nucleases (DHS1).Citation2-Citation5 In particular, nuc-1, the strictly positioned nucleosome immediately downstream of the LTR transcription start site (TSS), is highly repressive to transcription and specifically disrupted upon LTR activation.Citation3
Figure 1. (A) In vivo chromatin organization of the HIV LTR and distribution of putative transcription factor binding sites identified using the TF consite bioinformatics tool. (B) Location of the strictly positioned HIV-1 LTR nucleosomes correlates negatively with the predicted histone binding affinity score (nucleosome score) of the DNA sequence encompassing the HIV LTR. Predicted nucleosome affinity for HIV nucleotide sequence 1–720 was determinedCitation5 using the algorithm described.Citation25 Means and standard deviations for nucleosome score at insertion sites are indicated by dashed black (mean) and gray (SD) lines and give reference to known genomic sites of HIV integration.Citation63
Given the tight regulatory role played by its chromatin organization, transcription at the HIV LTR depends on host cell transcription factors and chromatin modifying activities to both promote the establishment of latency and to mediate LTR activation. Basal transcription from the HIV-1 promoter in the immediate early phase of HIV infection is stochastic and influenced by factors such as differences in the local chromatin microenvironment at the viral integration site and cellular transcription factors regulating transcription from the LTR.Citation6-Citation9 In this critical transcriptional phase, the intricate balance between the LTR-bound activating and repressive host factors, in large part determined by the chromatin environment of the integration site, will determine the fate of the integrated virus to a latent or transcriptionally active state.
The latent HIV-1 LTR is characterized by hypoacetylation of histones, resulting from recruitment of histone deacetylase enzymes (HDACs) by a number of repressive LTR-bound transcription factorsCitation10-Citation15 and increased trimethylation at histone 3 lysine 27Citation16,Citation17 and lysine 9.Citation18,Citation19 CpG methylation at the HIV LTR was shown to be another important mechanism, which contributes to maintenance of the latent transcriptional state.Citation20,Citation21
Productive viral replication is dependent on a shift from basal transcription to activated transcription, which occurs by the subsequent accumulation of viral Tat protein, a potent transactivator. In the absence of Tat expression, transcription initiation at the HIV LTR occurs normally; however, due to RNAP II pausing, only short abortive transcripts are produced. When expressed, Tat binds to TAR, an RNA stem-loop in the nascent viral RNA, and mediates transactivation at the LTR in at least two ways: Tat recruits the positive transcription elongation factor complex (pTEFb) containing cyclin T1 and CDK9, which phosphorylates the carboxyl-terminal domain of RNAP II leading to efficient elongation of transcription.Citation22,Citation23 Tat also orchestrates the recruitment of a number of chromatin modifying and remodeling complexes to the LTR, leading to extensive post-translational modifications of both LTR chromatin and Tat itself.Citation22,Citation23 In turn, more efficient transcription of the HIV genome, including Tat, generates a Tat-dependent positive feedback loop. Thus, we have a very detailed picture of the chromatin modifications and the chromatin modifying and remodeling activities associated with the HIV LTR in the transcriptionally repressed and activated states.
Predicted Vs. In Vivo LTR Nucleosome Structure
From a mechanistic perspective, the well characterized and strict chromatin organization of the HIV LTR, together with the presence of multiple transcription factor consensus sequences within the DHS1 and nuc-1 regions (), makes the HIV LTR an excellent model system to investigate the role of chromatin structure in transcription regulation. However, our understanding of the structural conformation of the HIV LTR and specifically the dynamic changes in chromatin architecture that result from LTR activation remains incompletely understood. This is highlighted by discrepancy in the literature between the in vivo and in vitro determined chromatin architecture of the LTR DHS1, which harbors host transcription factor binding sites critical for virus function. While the in vivo data shows this region to be sensitive to DNase digestion, suggesting it is devoid of nucleosomes,Citation2-Citation5 in vitro, DHS1 is assembled into a ternary complex of transcription factors, histones and DNA, suggesting the presence of a nucleosome.Citation24
We recently determined the predicted nucleosome affinity of the HIV LTR sequence using NuPoP software toolCitation25 and compared the predicted nucleosome distribution to the known in vivo LTR nucleosome positioning. This comparison demonstrated a striking reverse correlation ()Citation5: the DNA sequence encompassing the DHS1 region displayed the highest affinity for nucleosome formation while the strictly positioned nuc-0 and nuc-1 sequences displayed lower nucleosome propensity.Citation5 Thus, within the HIV LTR, the nucleosomes are not deposited according to their thermodynamically most favorable positions.
Our investigation of the HIV nucleosomal landscape using high resolution MNase mappingCitation5 also demonstrated that the DHS1 region of the HIV LTR, despite its high sequence propensity for nucleosome formation () contains unstable or loosely positioned nucleosomes.Citation5 This region of the LTR contains a tightly clustered distribution of putative binding sites for various transcription factors (depicted in ) identified using the TF consite bioinformatics tool, many of which have previously been shown to play crucial roles in LTR transcription regulation.Citation26
Early studies using in vitro assembled or positioned nucleosomes and purified transcription factors have shown that on the HIV LTR, binding of transcription factors to their recognition sites within nucleosomes is cooperative and context-dependent.Citation27-Citation31 For example, simultaneous but not individual binding of transcription factors to nucleosomes could dramatically loosen histone tail-DNA interactions at the LTRCitation31 and synergistically activated LTR-directed transcription on chromatin templates.Citation27 In this context, the HIV LTR can be regarded as an enhanceosome, formed by the cooperative assembly of multiple transcription factors, some of which open up chromatin or recruit nucleosome destabilizing cofactor complexes. The clustered, sometimes overlapping pattern of putative transcription factor binding sites include those of both activating and repressive transcription factors and cofactor complexes. This pattern of LTR transcription factor binding site distribution is consistent with a mechanism whereby competition between functionally opposing complexes on the same element leads to either productive or latent infections and underlies the stochastic nature of transcription at the LTR.
The concerted effect of these various binding sites and their relative orientations mediate combinatorial interactions between multiple DNA-transcription factor and co-factor complexes resulting in a structure thought to be inconsistent with the coexistence of stable nucleosomes. In agreement, we found that the LTR DHS1 region in its latent state is neither devoid of nucleosomes as previously assumed, nor assembled into a positioned nucleosome as predicted by its underlying sequence. Rather, DHS1 contains a loosely positioned nucleosome, which is evicted upon activation.Citation5 A recent genome wide study revealed that DHS sites, which are often assumed to be devoid of nucleosomes, are in fact occupied by histone variants H2AZ and H3.3.Citation32 In this regard, it would be interesting to examine the deposition of histone variants throughout the HIV LTR and elucidate their possible regulatory role in LTR activation.
SWI/SNF And The HIV Life Cycle
Nearly two decades ago, human SNF5/ Integrase Interactor-1 (INI-1), a core subunit to all mammalian ATP dependent SWI/SNF chromatin remodeling complexes, was first identified in a yeast two hybrid screen as a specific interactor of the HIV protein Integrase (IN).Citation33 Since then, studies have described different roles for INI-1/hSNF5 and the SWI/SNF complex in distinct steps of the HIV life cycle.
During HIV infection, incoming retroviral pre-integration complexes trigger the cytoplasmic export of the SWI/SNF component INI-1 and of the nuclear body constituent PML.Citation34 The HIV genome associates with these proteins before nuclear migration. In the presence of arsenic, PML is sequestered in the nucleus and the INI-1/pre-integration complex interaction is disrupted.Citation34 Under these conditions, the efficiency of HIV-mediated transduction is markedly increased, suggesting a repressive role for INI-1/hSNF5 in HIV transcription. INI-1/hSNF5 was shown to also inhibit early steps of HIV-1 replication by interacting with the HIV IN.Citation35 Recently, the interaction between the HIV IN and SWI/SNF was shown to facilitate virus integration into stable nucleosomes, functionally coupling the processes of virus integration and remodeling.Citation36 Moreover, INI-1/hSNF5 was suggested to play an additional role in the HIV life cycle, to facilitate virus assembly or release.Citation37
In addition to the non-transcriptional roles described for SWI/SNF in the HIV life cycle, others and we found INI-1 and SWI/SNF to be directly involved in Tat-dependent activation of transcription at the HIV LTR.Citation5,Citation38-Citation42 SWI/SNF was shown to also mediate Tat-independent transcription elongation at the HIV LTR.Citation43 Furthermore, in addition to its direct involvement in LTR activation, SWI/SNF was shown to repress basal LTR activity.Citation5,Citation44 Thus, a complex picture emerges from the regulatory role of SWI/SNF and its core subunit INI-1 in the various stages of the HIV-1 life cycle, from virus integration and LTR transcription to replication and viral assembly. Importantly, the seemingly contradictory role of SWI/SNF in both transcription activation and repression at the LTR highlighted our incomplete understanding of the mechanisms behind SWI/SNF regulation of LTR transcription.
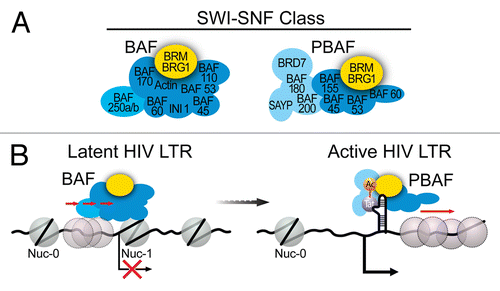
At least two biochemically distinct SWI/SNF sub-complexes exist, the BAF and the PBAF complexes, which appear to have different functions. The PBAF complex contains either BRG1 or BRM together with the PBAF-specific subunits BAF180, BAF200, SAYP and Brd7, but lacks BAF250.Citation45-Citation49 The BAF complex contains either BRG1 or BRM together with the BAF-specific subunit BAF250, but lacks PBAF-specific subunitsCitation50,Citation51 (). The presence of the biochemically and functionally distinct SWI/SNF sub-complexes BAF and PBAF () has been critical to distinguish between SWI/SNF-mediated LTR repression and Tat-mediated SWI/SNF recruitment. The presence of unique subunits in the distinct BAF and PBAF complexes has also been instrumental to elucidate the underlying mechanism of LTR transcription regulation mediated by each SWI/SNF sub-complex. We found recently that repression at the HIV LTR is an active process driven by ATP hydrolysis. The distinct BAF complex specifically functions to counteract intrinsic histone-DNA sequence preferences at the LTR to move a preferred nucleosome from DHS1 to position nuc-1 over sub-optimal sequences immediately downstream of the TSS.Citation5 Thus, BAF is required for LTR repression and maintenance of latency.Citation5 Upon activation, BAF dissociates from the LTR, and the distinct PBAF complex is recruited by Tat to facilitate transcriptionCitation5,Citation42 (). Thus, the distinct BAF and PBAF complexes perform transcriptionally opposing functions in regulating LTR activity.
Figure 2. (A) Subunit composition of the two distinct mammalian SWI/SNF complexes, BAF and PBAF. (B) Model for SWI/SNF regulation of HIV LTR transcription. BAF actively counteracts intrinsic histone-DNA sequence preferences within HIV LTR and pulls a preferred nucleosome over DHS1 onto DNA sequences less favorable for nucleosome formation immediately downstream of the TSS, leading to positioning of nuc-1 and transcriptional repression. Upon activation, BAF dissociates from the LTR resulting in de-repression of HIV transcription and expression of Tat. Acetylated Tat selectively recruits the PBAF complex, which actively re-positions nucleosomes formed downstream of TSS enabling efficient transcription elongation.
LTR Targeting Of BAF
An important question remaining to be resolved is how BAF is recruited to the HIV LTR to position the repressive nuc-1. SWI/SNF remodelers have been shown to bind to distinct genomic loci.Citation52,Citation53 There are three nonexclusive mechanisms that have been described for targeting of SWI/SNF to its specific sites. SWI/SNF complexes have been shown to be recruited via interaction with sequence-specific transcription factors by recognition of a particular combination of histone modifications or, directly, by binding to a specific DNA structure.Citation54-Citation56
The HIV LTR contains binding sites for multiple sequence-specific host transcription factors (). One possible mechanism is that a nuc-1 associated repressive transcription factor recruits BAF to the HIV LTR. A number of transcriptional repressors contain binding sites within the LTR DHS1 and nuc-1 regions, including LBP-1 and YY-1.Citation10 Indeed, YY-1 was bound to the HIV promoter under basal conditions and displaced in response to Tat expression.Citation39 Thus, YY-1 is a candidate transcription factor, which may recruit BAF to the HIV LTR to position the repressive nuc-1. In support of this possibility, the Drosophila homolog of YY-1, pleiohomeotic (PHO), directly binds Brahma, the Drosophila SWI/SNF complex, recruiting it to target genes.Citation57 In addition, other LTR-bound transcription factors such as AP-1 and NFAT have been shown previously to bind and recruit SWI/SNF to target genesCitation58,Citation59 and may contribute to BAF recruitment at the LTR. Other factors such as the presence of a distinct pattern of histone modifications over the LTR, or a unique DNA structure within the LTR, may also contribute to the specific LTR recruitment of BAF.
BAF And LTR Nucleosome Positioning
ATP dependent chromatin remodeling complexes are considered to regulate gene expression by using energy from ATP hydrolysis to create nucleosome-depleted regions by disrupting already positioned nucleosomes.Citation60 These complexes contribute to the ejection and disruption of positioned nucleosomes from enhancer/promoter regions and remodel or mobilize nucleosomes along the flanking chromatin. This model implies that, in vivo, nucleosomes assemble and distribute according to their underlying histone-DNA sequence preferences at intrinsically defined positions.Citation60 However, recently, a number of studies have presented compelling evidence to suggest that ATP-dependent remodeling complexes effect nucleosome formation, on a global scale, by actively countering the preferred positions dictated by underlying DNA sequences.Citation53,Citation61,Citation62 Genome-wide studies in yeast probed the effect of ATP depletionCitation61 as well as the specific depletion of ISWI class chromatin remodelersCitation62 on nucleosome positioning. Another seminal study examined the effect of depletion of remodelers of different classes on global nucleosome positioning in the Drosophila genome.Citation53 These studies demonstrated that ATP-dependent chromatin remodelers actively counteract DNA sequence preferred nucleosome distribution. Of particular interest, Drosophila SWI/SNF or (P)BAP was shown to increase nucleosome density at its target sequences or, in other words, to “pull” nucleosomes over unfavorable DNA sequences.Citation53 In agreement to what is observed on a global genomic scale, our data on the HIV LTR indicates that BAF counters the intrinsic DNA-driven nucleosome placement preference conferred by the LTR sequence to generate an LTR chromatin structure that is repressive to transcription.Citation5
Why would the HIV virus position a repressive nucleosome immediately downstream of the TSS? It is possible that BAF pulls the thermodynamically favored nucleosome away from the DHS1, which contains binding sites for host cell transcription factors, in order to allow for the assembly of the transcription initiation complex. In support of this model, a previous study using in vitro reconstituted and positioned LTR DHS1 argued for the presence of a ternary complex consisting of transcription factors, histones, and DNA over the DHS1 region in vitro.Citation24 Our modeling data, depicting the high predicted affinity (nucleosome score) of the DHS1 DNA sequence () is in agreement with these observations. In addition, when BAF was depleted by siRNA in latent HIV infected Jurkat cells, DNA accessibility decreased over DHS1 concomitant with an increase in histone density, arguing for the presence of a complex over DHS1 consisting of a nucleosome, possibly together with transcription factors.Citation5 Together, these observations support a regulatory model in which BAF actively pulls a preferred nucleosome from DHS1 to allow for binding and assembly of the transcription initiation complex, and positions a nucleosome over unfavorable sequences immediately downstream of the TSS to repress transcription. Why position a repressive nucleosome over a DNA sequence that is intrinsically less favorable for positioning? A key aspect of transcription regulation is likely to be the maintenance of positioned nucleosomes over regulatory sequences until the appropriate signals are received to permit their destabilization. From a regulatory perspective, this unfavorable positioning might facilitate the signal dependent remodeling of such a thermodynamically sub-optimal nucleosome. Thus, on the HIV LTR, the antagonism between ATP-driven BAF action and DNA sequence preference effects on nucleosome positioning may allow for a speedy de-repression of transcription in response to the appropriate signals.
BAF And HIV Latency
The identification of different molecular mechanisms targeting different pathways involved in HIV LTR transcriptional silencing will contribute to define a combinatorial strategy to activate latent HIV, an approach that in combination with c-ART could lead to curative therapies. The energetically unfavorable positioning of the repressive LTR nuc-1 by the BAF complex provides a novel molecular mechanism for latency and an added level of complexity to the mechanisms that work together to transcriptionally silence the HIV LTR. In this context, the enzyme BRG1, as the ATP-dependent catalytic subunit of the repressive BAF complex may be an attractive therapeutic target for small molecule inhibition in depletion of the latent reservoir from HIV infected patients.
Acknowledgments
I thank Yuri Moshkin for helpful discussions and Haleh Rafati for graphical assistance. This work was supported by an ErasmusMC Fellowship.
References
- Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. The challenge of finding a cure for HIV infection. Science 2009; 323:1304 - 7; http://dx.doi.org/10.1126/science.1165706; PMID: 19265012
- Verdin E. DNase I-hypersensitive sites are associated with both long terminal repeats and with the intragenic enhancer of integrated human immunodeficiency virus type 1. J Virol 1991; 65:6790 - 9; PMID: 1942252
- Verdin E, Paras PJ Jr., Van Lint C. Chromatin disruption in the promoter of human immunodeficiency virus type 1 during transcriptional activation. EMBO J 1993; 12:3249 - 59; PMID: 8344262
- Van Lint C, Emiliani S, Ott M, Verdin E. Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J 1996; 15:1112 - 20; PMID: 8605881
- Rafati H, Parra M, Hakre S, Moshkin Y, Verdin E, Mahmoudi T. Repressive LTR nucleosome positioning by the BAF complex is required for HIV latency. PLoS Biol 2011; 9:e1001206; http://dx.doi.org/10.1371/journal.pbio.1001206; PMID: 22140357
- Jordan A, Defechereux P, Verdin E. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J 2001; 20:1726 - 38; http://dx.doi.org/10.1093/emboj/20.7.1726; PMID: 11285236
- Schröder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 2002; 110:521 - 9; http://dx.doi.org/10.1016/S0092-8674(02)00864-4; PMID: 12202041
- Weinberger LS, Dar RD, Simpson ML. Transient-mediated fate determination in a transcriptional circuit of HIV. Nat Genet 2008; 40:466 - 70; http://dx.doi.org/10.1038/ng.116; PMID: 18344999
- Burnett JC, Miller-Jensen K, Shah PS, Arkin AP, Schaffer DV. Control of stochastic gene expression by host factors at the HIV promoter. PLoS Pathog 2009; 5:e1000260; http://dx.doi.org/10.1371/journal.ppat.1000260; PMID: 19132086
- Coull JJ, Romerio F, Sun JM, Volker JL, Galvin KM, Davie JR, et al. The human factors YY1 and LSF repress the human immunodeficiency virus type 1 long terminal repeat via recruitment of histone deacetylase 1. J Virol 2000; 74:6790 - 9; http://dx.doi.org/10.1128/JVI.74.15.6790-6799.2000; PMID: 10888618
- Tyagi M, Karn J. CBF-1 promotes transcriptional silencing during the establishment of HIV-1 latency. EMBO J 2007; 26:4985 - 95; http://dx.doi.org/10.1038/sj.emboj.7601928; PMID: 18007589
- Williams SA, Chen LF, Kwon H, Ruiz-Jarabo CM, Verdin E, Greene WC. NF-kappaB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J 2006; 25:139 - 49; http://dx.doi.org/10.1038/sj.emboj.7600900; PMID: 16319923
- Jiang G, Espeseth A, Hazuda DJ, Margolis DM. c-Myc and Sp1 contribute to proviral latency by recruiting histone deacetylase 1 to the human immunodeficiency virus type 1 promoter. J Virol 2007; 81:10914 - 23; http://dx.doi.org/10.1128/JVI.01208-07; PMID: 17670825
- Marban C, Suzanne S, Dequiedt F, de Walque S, Redel L, Van Lint C, et al. Recruitment of chromatin-modifying enzymes by CTIP2 promotes HIV-1 transcriptional silencing. EMBO J 2007; 26:412 - 23; http://dx.doi.org/10.1038/sj.emboj.7601516; PMID: 17245431
- Imai K, Okamoto T. Transcriptional repression of human immunodeficiency virus type 1 by AP-4. J Biol Chem 2006; 281:12495 - 505; http://dx.doi.org/10.1074/jbc.M511773200; PMID: 16540471
- Friedman J, Cho WK, Chu CK, Keedy KS, Archin NM, Margolis DM, et al. Epigenetic silencing of HIV-1 by the histone H3 lysine 27 methyltransferase enhancer of Zeste 2. J Virol 2011; 85:9078 - 89; http://dx.doi.org/10.1128/JVI.00836-11; PMID: 21715480
- Kim HG, Kim KC, Roh TY, Park J, Jung KM, Lee JS, et al. Gene silencing in HIV-1 latency by polycomb repressive group. Virol J 2011; 8:179; http://dx.doi.org/10.1186/1743-422X-8-179; PMID: 21496352
- du Chéné I, Basyuk E, Lin YL, Triboulet R, Knezevich A, Chable-Bessia C, et al. Suv39H1 and HP1gamma are responsible for chromatin-mediated HIV-1 transcriptional silencing and post-integration latency. EMBO J 2007; 26:424 - 35; http://dx.doi.org/10.1038/sj.emboj.7601517; PMID: 17245432
- Mateescu B, Bourachot B, Rachez C, Ogryzko V, Muchardt C. Regulation of an inducible promoter by an HP1beta-HP1gamma switch. EMBO Rep 2008; 9:267 - 72; http://dx.doi.org/10.1038/embor.2008.1; PMID: 18239689
- Kauder SE, Bosque A, Lindqvist A, Planelles V, Verdin E. Epigenetic regulation of HIV-1 latency by cytosine methylation. PLoS Pathog 2009; 5:e1000495; http://dx.doi.org/10.1371/journal.ppat.1000495; PMID: 19557157
- Blazkova J, Trejbalova K, Gondois-Rey F, Halfon P, Philibert P, Guiguen A, et al. CpG methylation controls reactivation of HIV from latency. PLoS Pathog 2009; 5:e1000554; http://dx.doi.org/10.1371/journal.ppat.1000554; PMID: 19696893
- Easley R, Van Duyne R, Coley W, Guendel I, Dadgar S, Kehn-Hall K, et al. Chromatin dynamics associated with HIV-1 Tat-activated transcription. Biochim Biophys Acta 2010; 1799:275 - 85; PMID: 19716452
- Karn J, Stoltzfus CM. Transcriptional and Posttranscriptional Regulation of HIV-1 Gene Expression. Cold Spring Harb Perspect Med 2012; 2:a006916; PMID: 22355797
- Steger DJ, Workman JL. Stable co-occupancy of transcription factors and histones at the HIV-1 enhancer. EMBO J 1997; 16:2463 - 72; http://dx.doi.org/10.1093/emboj/16.9.2463; PMID: 9171359
- Xi L, Fondufe-Mittendorf Y, Xia L, Flatow J, Widom J, Wang JP. Predicting nucleosome positioning using a duration Hidden Markov Model. BMC Bioinformatics 2010; 11:346; http://dx.doi.org/10.1186/1471-2105-11-346; PMID: 20576140
- Pereira LA, Bentley K, Peeters A, Churchill MJ, Deacon NJ. A compilation of cellular transcription factor interactions with the HIV-1 LTR promoter. Nucleic Acids Res 2000; 28:663 - 8; http://dx.doi.org/10.1093/nar/28.3.663; PMID: 10637316
- Pazin MJ, Sheridan PL, Cannon K, Cao Z, Keck JG, Kadonaga JT, et al. NF-kappa B-mediated chromatin reconfiguration and transcriptional activation of the HIV-1 enhancer in vitro. Genes Dev 1996; 10:37 - 49; http://dx.doi.org/10.1101/gad.10.1.37; PMID: 8557193
- Sheridan PL, Mayall TP, Verdin E, Jones KA. Histone acetyltransferases regulate HIV-1 enhancer activity in vitro. Genes Dev 1997; 11:3327 - 40; http://dx.doi.org/10.1101/gad.11.24.3327; PMID: 9407026
- Widlak P, Gaynor RB, Garrard WT. In vitro chromatin assembly of the HIV-1 promoter. ATP-dependent polar repositioning of nucleosomes by Sp1 and NFkappaB. J Biol Chem 1997; 272:17654 - 61; http://dx.doi.org/10.1074/jbc.272.28.17654; PMID: 9211915
- Steger DJ, Eberharter A, John S, Grant PA, Workman JL. Purified histone acetyltransferase complexes stimulate HIV-1 transcription from preassembled nucleosomal arrays. Proc Natl Acad Sci U S A 1998; 95:12924 - 9; http://dx.doi.org/10.1073/pnas.95.22.12924; PMID: 9789016
- Angelov D, Charra M, Seve M, Côté J, Khochbin S, Dimitrov S. Differential remodeling of the HIV-1 nucleosome upon transcription activators and SWI/SNF complex binding. J Mol Biol 2000; 302:315 - 26; http://dx.doi.org/10.1006/jmbi.2000.4069; PMID: 10970736
- Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, et al. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet 2009; 41:941 - 5; http://dx.doi.org/10.1038/ng.409; PMID: 19633671
- Kalpana GV, Marmon S, Wang W, Crabtree GR, Goff SP. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science 1994; 266:2002 - 6; http://dx.doi.org/10.1126/science.7801128; PMID: 7801128
- Turelli P, Doucas V, Craig E, Mangeat B, Klages N, Evans R, et al. Cytoplasmic recruitment of INI1 and PML on incoming HIV preintegration complexes: interference with early steps of viral replication. Mol Cell 2001; 7:1245 - 54; http://dx.doi.org/10.1016/S1097-2765(01)00255-6; PMID: 11430827
- Maroun M, Delelis O, Coadou G, Bader T, Ségéral E, Mbemba G, et al. Inhibition of early steps of HIV-1 replication by SNF5/Ini1. J Biol Chem 2006; 281:22736 - 43; http://dx.doi.org/10.1074/jbc.M604849200; PMID: 16772295
- Lesbats P, Botbol Y, Chevereau G, Vaillant C, Calmels C, Arneodo A, et al. Functional coupling between HIV-1 integrase and the SWI/SNF chromatin remodeling complex for efficient in vitro integration into stable nucleosomes. PLoS Pathog 2011; 7:e1001280; http://dx.doi.org/10.1371/journal.ppat.1001280; PMID: 21347347
- Yung E, Sorin M, Pal A, Craig E, Morozov A, Delattre O, et al. Inhibition of HIV-1 virion production by a transdominant mutant of integrase interactor 1. Nat Med 2001; 7:920 - 6; http://dx.doi.org/10.1038/90959; PMID: 11479624
- Tréand C, du Chéné I, Brès V, Kiernan R, Benarous R, Benkirane M, et al. Requirement for SWI/SNF chromatin-remodeling complex in Tat-mediated activation of the HIV-1 promoter. EMBO J 2006; 25:1690 - 9; http://dx.doi.org/10.1038/sj.emboj.7601074; PMID: 16601680
- Mahmoudi T, Parra M, Vries RG, Kauder SE, Verrijzer CP, Ott M, et al. The SWI/SNF chromatin-remodeling complex is a cofactor for Tat transactivation of the HIV promoter. J Biol Chem 2006; 281:19960 - 8; http://dx.doi.org/10.1074/jbc.M603336200; PMID: 16687403
- Agbottah E, Deng L, Dannenberg LO, Pumfery A, Kashanchi F. Effect of SWI/SNF chromatin remodeling complex on HIV-1 Tat activated transcription. Retrovirology 2006; 3:48; http://dx.doi.org/10.1186/1742-4690-3-48; PMID: 16893449
- Ariumi Y, Serhan F, Turelli P, Telenti A, Trono D. The integrase interactor 1 (INI1) proteins facilitate Tat-mediated human immunodeficiency virus type 1 transcription. Retrovirology 2006; 3:47; http://dx.doi.org/10.1186/1742-4690-3-47; PMID: 16889668
- Easley R, Carpio L, Dannenberg L, Choi S, Alani D, Van Duyne R, et al. Transcription through the HIV-1 nucleosomes: effects of the PBAF complex in Tat activated transcription. Virology 2010; 405:322 - 33; http://dx.doi.org/10.1016/j.virol.2010.06.009; PMID: 20599239
- Mizutani T, Ishizaka A, Tomizawa M, Okazaki T, Yamamichi N, Kawana-Tachikawa A, et al. Loss of the Brm-type SWI/SNF chromatin remodeling complex is a strong barrier to the Tat-independent transcriptional elongation of human immunodeficiency virus type 1 transcripts. J Virol 2009; 83:11569 - 80; http://dx.doi.org/10.1128/JVI.00742-09; PMID: 19726504
- Boese A, Sommer P, Holzer D, Maier R, Nehrbass U. Integrase interactor 1 (Ini1/hSNF5) is a repressor of basal human immunodeficiency virus type 1 promoter activity. J Gen Virol 2009; 90:2503 - 12; http://dx.doi.org/10.1099/vir.0.013656-0; PMID: 19515827
- Xue Y, Canman JC, Lee CS, Nie Z, Yang D, Moreno GT, et al. The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proc Natl Acad Sci U S A 2000; 97:13015 - 20; http://dx.doi.org/10.1073/pnas.240208597; PMID: 11078522
- Lemon B, Inouye C, King DS, Tjian R. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature 2001; 414:924 - 8; http://dx.doi.org/10.1038/414924a; PMID: 11780067
- Chalkley GE, Moshkin YM, Langenberg K, Bezstarosti K, Blastyak A, Gyurkovics H, et al. The transcriptional coactivator SAYP is a trithorax group signature subunit of the PBAP chromatin remodeling complex. Mol Cell Biol 2008; 28:2920 - 9; http://dx.doi.org/10.1128/MCB.02217-07; PMID: 18299390
- Yan Z, Cui K, Murray DM, Ling C, Xue Y, Gerstein A, et al. PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev 2005; 19:1662 - 7; http://dx.doi.org/10.1101/gad.1323805; PMID: 15985610
- Kaeser MD, Aslanian A, Dong MQ, Yates JR 3rd, Emerson BM. BRD7, a novel PBAF-specific SWI/SNF subunit, is required for target gene activation and repression in embryonic stem cells. J Biol Chem 2008; 283:32254 - 63; http://dx.doi.org/10.1074/jbc.M806061200; PMID: 18809673
- Nie Z, Xue Y, Yang D, Zhou S, Deroo BJ, Archer TK, et al. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol Cell Biol 2000; 20:8879 - 88; http://dx.doi.org/10.1128/MCB.20.23.8879-8888.2000; PMID: 11073988
- Nagl NG Jr., Wang X, Patsialou A, Van Scoy M, Moran E. Distinct mammalian SWI/SNF chromatin remodeling complexes with opposing roles in cell-cycle control. EMBO J 2007; 26:752 - 63; http://dx.doi.org/10.1038/sj.emboj.7601541; PMID: 17255939
- Ng HH, Robert F, Young RA, Struhl K. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev 2002; 16:806 - 19; http://dx.doi.org/10.1101/gad.978902; PMID: 11937489
- Moshkin YM, Chalkley GE, Kan TW, Reddy BA, Ozgur Z, van Ijcken WF, et al. Remodelers organize cellular chromatin by counteracting intrinsic histone-DNA sequence preferences in a class-specific manner. Mol Cell Biol 2012; 32:675 - 88; http://dx.doi.org/10.1128/MCB.06365-11; PMID: 22124157
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem 2009; 78:273 - 304; http://dx.doi.org/10.1146/annurev.biochem.77.062706.153223; PMID: 19355820
- Ho L, Crabtree GR. Chromatin remodelling during development. Nature 2010; 463:474 - 84; http://dx.doi.org/10.1038/nature08911; PMID: 20110991
- Quinn J, Fyrberg AM, Ganster RW, Schmidt MC, Peterson CL. DNA-binding properties of the yeast SWI/SNF complex. Nature 1996; 379:844 - 7; http://dx.doi.org/10.1038/379844a0; PMID: 8587611
- Mohd-Sarip A, Venturini F, Chalkley GE, Verrijzer CP. Pleiohomeotic can link polycomb to DNA and mediate transcriptional repression. Mol Cell Biol 2002; 22:7473 - 83; http://dx.doi.org/10.1128/MCB.22.21.7473-7483.2002; PMID: 12370294
- Ito T, Yamauchi M, Nishina M, Yamamichi N, Mizutani T, Ui M, et al. Identification of SWI.SNF complex subunit BAF60a as a determinant of the transactivation potential of Fos/Jun dimers. J Biol Chem 2001; 276:2852 - 7; http://dx.doi.org/10.1074/jbc.M009633200; PMID: 11053448
- Wurster AL, Pazin MJ. BRG1-mediated chromatin remodeling regulates differentiation and gene expression of T helper cells. Mol Cell Biol 2008; 28:7274 - 85; http://dx.doi.org/10.1128/MCB.00835-08; PMID: 18852284
- Cockerill PN. Structure and function of active chromatin and DNase I hypersensitive sites. FEBS J 2011; 278:2182 - 210; http://dx.doi.org/10.1111/j.1742-4658.2011.08128.x; PMID: 21501387
- Zhang Z, Wippo CJ, Wal M, Ward E, Korber P, Pugh BF. A packing mechanism for nucleosome organization reconstituted across a eukaryotic genome. Science 2011; 332:977 - 80; http://dx.doi.org/10.1126/science.1200508; PMID: 21596991
- Gkikopoulos T, Schofield P, Singh V, Pinskaya M, Mellor J, Smolle M, et al. A role for Snf2-related nucleosome-spacing enzymes in genome-wide nucleosome organization. Science 2011; 333:1758 - 60; http://dx.doi.org/10.1126/science.1206097; PMID: 21940898
- Brady T, Agosto LM, Malani N, Berry CC, O’Doherty U, Bushman F. HIV integration site distributions in resting and activated CD4+ T cells infected in culture. AIDS 2009; 23:1461 - 71; http://dx.doi.org/10.1097/QAD.0b013e32832caf28; PMID: 19550285