Abstract
TAF7, a component of the TFIID complex, controls the first steps of transcription. It interacts with and regulates the enzymatic activities of transcription factors that regulate RNA polymerase II progression. Its diverse functions in transcription initiation are consistent with its essential role in cell proliferation.
Introduction
Productive transcription of RNA polymerase II (RNAP II)-dependent genes requires an ordered series of events that lead to initiation, elongation and termination. Transcription initiation is preceded by the assembly of the RNAP II machinery into a preinitiation complex (PIC).Citation1 PIC assembly begins with the binding of the general transcription factor, TFIID, to the core promoter, followed by the sequential recruitment of the general transcription factors (GTFs) TFIIB, TFIIE, TFIIF and TFIIH. PIC assembly is completed by the association of Mediator, the bromodomain protein BRD4, the elongation factor P-TEFb and RNAP II.Citation1-Citation4 Once the PIC is fully assembled, transcription initiates with the incorporation of the first NTPs. The transition from initiation to productive elongation is accompanied by the phosphorylation of the C-terminal domain (CTD) of RNAP II by a series of kinases: CDK7/TFIIH, which phosphorylates Ser5 of the CTD heptad repeat, BRD4, which phosphorylates Ser2 during initiation and, finally, CDK9/PTEFb, which phosphorylates Ser5/2 during elongation.Citation5 These multiple phosphorylation events generate a platform on the CTD for the recruitment of multiple complexes necessary for the processing and maturation of nascent RNA.Citation5-Citation7 Whereas the sequence of events necessary for productive transcription initiation and elongation have been extensively characterized, little is known about the regulatory mechanisms that ensure that each step is successfully completed before the next one begins. Recent evidence indicates that the transcription factor, TAF7, plays a critical role in this regulation.
The TFIID complexes that bind to the core promoter are composed of either the TATA binding protein (TBP) or a TBP-related protein (TRF1, TRF2, TRF3) and over a dozen TBP Associated Factors (TAFs).Citation1,Citation8-Citation10 One of the TAFs found in the TFIID complex is TAF7, a 55 kD protein. Although TAF7 was originally thought to be solely a structural component of TFIID, work from our lab has established that it has a much broader role as a regulator of the activities of the various enzymes required for transcription initiation. The following sections summarize our current knowledge about TAF7, its interactions with other transcription factors and its role in regulating transcription initiation and cellular proliferation.
TAF7 regulates transcription initiation
Within TFIID, TAF7 is bound to TAF1. Among the TAFs, only the 250 kD TAF1 has enzymatic activities: two distinct kinase activities, mapping to the amino- and C-terminal domains, and an acetyltransferase activity.Citation11,Citation12 Substrates for the TAF1 kinase activities include TAF1 itself and TAF7.Citation11,Citation13 The intrinsic acetyl transferase (AT) activity of TAF1 is essential for transcription, both in vitro and in vivo, although its biological substrate has not been fully characterized. Despite the finding that the histone H3 associated with the cyclin D1 promoter is acetylated on K9 and K14 by TAF1,Citation14,Citation15 histones may not be the functionally relevant target of TAF1 AT activity. Indeed, TAF1 AT activity is required for transcription of naked DNA in vitro.Citation16 The interaction of TAF7 with TAF1 is through both its AT domain and the RAPiD domain, the region where the TFIIF RAP70 subunit also binds. In our early studies, we demonstrated that TAF7 binding to TAF1 inhibits its essential AT activity, thereby blocking transcription initiation. Exogenous TAF7 repressed expression of a TAF1-dependent gene, an MHC class I gene, both in vitro and in vivo.Citation17 These findings were confirmed and extended in a recent report that TAF7 overexpression represses TAF1 AT activity and inhibits cyclin D and cyclin A gene expression.Citation18
The fact that TAF7 binding to TAF1 inhibits the AT activity that is required for transcription means that the canonical TFIID holocomplex is transcriptionally repressed. This raises the paradoxical question: how does transcription initiate if TFIID is inactive? There are two possibilities: i) TAF7 dissociates from TFIID prior to PIC assembly or ii) TAF7 remains associated with TFIID during PIC assembly but is released subsequently. We have shown that the latter is the case: TAF7 is released from the PIC upon completion of assembly and concomitant with initiation.Citation13 Thus, TAF7 acts as a negative regulator of the earliest step of transcription initiation and may prevent premature, abortive initiation until all components necessary for transcription are in place. A corollary to the finding that TAF7 is released from TFIID is that TFIID is a highly dynamic complex, rather than the static, architectural one that it had been considered previously.
Although TAF7 regulates TAF1-dependent promoters, not all transcription is TAF1 dependent. For example, in the HEK293 kidney fibroblast line, only 54% of the transcripts require TAF1. Surprisingly, in the same cells, 65% of the transcripts depend on TAF7.Citation19 Thus, there is a subset of genes that are TAF7-dependent but TAF1-independent. (A similar disparity has been reported in yeast, where TAF7 regulates 27% of the transcripts whereas TAF1 is required for only 14%).Citation20 This leads to the question of what TAF7 targets in TAF1-independent transcription. One of the best characterized examples of TAF1-independent transcription is that of ϒ-interferon-activated transcription of MHC class I genes. Basal MHC class I gene transcription is TAF1 dependent. However, following ϒ-interferon (IFN) treatment of cells, class I transcription becomes independent of TAF1. Its functions are replaced by the IFN-induced transcriptional co-activator, CIITA. CIITA, like TAF1, has AT activity. It also interacts with TBP, TAF6 and TAF9,Citation19 suggesting that CIITA may form a TFIID-like structure. These characteristics allow CIITA to bypass the requirement for TAF1.Citation21 Importantly, TAF7 binds to CIITA inhibiting its AT activity and repressing transcription. Thus, TAF7 regulation of transcription initiation extends beyond its effect on TFIID.
Interestingly, of the TAF1-dependent genes, 31% are TAF7 independent. What regulates transcription initiation of these genes remains to be investigated.
TAF7 regulates the major RNAP II CTD kinases
In addition to its regulation of TAF1, TAF7 also interacts with, regulates and is regulated by the kinases that phosphorylated the RNAP II CTD during the early steps of transcription. The CTD consists of repeats of the heptamer Y1S2P3T4S5P6S7—the number of repeats varies in different organisms (52 in mammals, 26 in yeast). Phosphorylation of CTD Ser5/7, Ser2 and Ser5/2 residues by CDK7/TFIIH, BRD4 and CDK9/PTEFb, respectively, is necessary for the recruitment of different complexes involved in nascent RNA processing. The phosphorylation of Ser5 by CDK7/TFIIH occurs at the earliest step of transcription initiation when RNAP II is still at the promoter and concomitant with the synthesis of the first oligonucleotides. Ser2 is phosphorylated by BRD4 prior to the transition to efficient elongation. Subsequent Ser5/2 phosphorylation during elongation is mediated by CDK9/PTEFb.Citation5,Citation6
Surprisingly, TAF7 binds to and inhibits all three CTD kinases.Citation5,Citation22 TAF7 inhibition is specific both for these kinases and the CTD substrate. It does not inhibit CDK2, a minor CTD kinase, but it enhances PTEFb phosphorylation of SPT5, a component of the DSIF complex that switches from a negative regulator to a positive one early in transcription.Citation22,Citation23 While the activities of the CTD kinases are regulated by TAF7 binding, TAF7 is also phosphorylated by the three CTD kinases: CDK7 and BRD4 phosphorylate TAF7 in its C-terminal domain, whereas P-TEFb phosphorylates TAF7 in its central domain (unpublished data).Citation13,Citation24 Furthermore, TAF7 also is phosphorylated by TAF1, which results in the release of TAF7 from TFIID upon transcription initiation.Citation13,Citation18 Although TAF7 is phosphorylated at multiple sites by TAF1, recent evidence suggests that TAF7 Ser264 is the major site.Citation18 The phosphorylation of TAF7 by each of these kinases modulates its subsequent ability to inhibit the other CTD kinases, thereby creating a regulatory cascade that ensures an orderly progression through the steps of transcription initiation.Citation24 Based on these observations, we have proposed that TAF7 plays a key role in regulating transcription, functioning as a check point regulator that modulates the enzymatic activities required for initiation and elongation of transcription, ensuring that each step in the process is completed before continuing to the next one.Citation13,Citation22
As noted above, TAF7 regulates transcription of both TAF1-dependent and -independent genes. The general mechanism(s) by which TAF7 regulates TAF1-independent genes remains to be fully characterized. However, the interaction of TAF7 with the major RNA polymerase II CTD kinases provides one possible regulatory mechanism.
TAF7-dependent transcripts are essential for cell proliferation
As noted above, not all transcription is TAF7 dependent. T cell maturation in the thymus and the periphery provided an ideal model system for us to examine whether there was a correlation between TAF7-dependent transcription and proliferation or differentiation. The stages of thymocyte differentiation are distinguished by their expression of two cell surface co-receptor molecules CD4 and CD8. The most immature thymic precursors express neither CD4 nor CD8 (double negative thymocytes, DN). Following rearrangement of the T cell receptor β gene (TCRβ), DN thymocytes undergo massive proliferation and differentiate into “double positive” (DP) thymocytes that express both CD4 and CD8. Subsequent differentiation of DP thymocytes to either mature single positive CD4 (CD4SP) or CD8 (CD8SP) thymocytes proceeds without further proliferation.Citation25
Deletion of TAF7 at different stages of thymocyte development has dramatically different effects. TAF7 deletion at the DN stage, which requires proliferation to progress to the DP stage, blocks cellular expansion and arrests further development. Proliferation, rather than survival, of DN cells is affected by TAF7 loss as evidenced by: i) TAF7-deleted DN thymocytes do not incorporate BrdU efficiently, relative to wild type DN cells and, ii) cell loss is not rescued by introduction of a transgene encoding the survival molecule, Bcl2 (unpublished data).Citation26 In marked contrast, deletion of TAF7 at the DP positive stage, which does not require proliferation to transition to the SP stage, has only a minor effect on thymocyte development. Indeed, TAF7-deleted DP thymocytes differentiate into CD4 and CD8 SP thymocytes, exit the thymus and populate peripheral lymphoid organs. TAF7-deleted T cells in peripheral lymphoid organs are unable to proliferate in response to activation stimuli but are capable of de novo transcription of cell surface markers diagnostic of the TCR activation pathway, such as CD69.Citation26 Thus TAF7 is not required for all transcription but is an absolute requirement for proliferation at all stages of T cell development.
Many other cell types also depend on TAF7, in toto or in part, for proliferation. TAF7 depletion in HEK293 kidney cellsCitation22 or in hamster ovary CHO cells (unpublished data) significantly retards proliferation. Mouse embryonic fibroblasts (MEFs) stop proliferating following TAF7 deletion.Citation26 Germline deletion of TAF7 in mice is embryonic lethal between 4.5 and 5.5 d post coitus, the time at which the embryo implants and undergoes marked cellular expansion.Citation26 Taken together, these findings suggest that all cells have a requirement for TAF7 to proliferate.
Although TAF7 is necessary for proliferation, it is not required at a specific stage in the cell cycle. TAF7−/− MEFs are blocked throughout cell cycle.Citation26 Furthermore, overexpression of TAF7 in MEFs also does not cause a specific cell cycle arrest (unpublished data). Thus, although TAF7 interacts with TAF1 and BRD4, mutation or deletion of either causes an arrest in cell cycle at the G1/S interface, TAF7 deletion does not have a similar effect.Citation26-Citation28
Consistent with its critical role in proliferation, transcription in immature, proliferating cells is TAF7-dependent. But this requirement diminishes during differentiation. Thus, deletion of TAF7 from proliferating MEFs results in a global repression of transcription, with nearly all genes affected.Citation26 This is not due to a defect in the TFIID complex: the holo-TFIID complex continues to be assembled in these cells despite the absence of TAF7. In the more differentiated but still proliferative HEK293 kidney cell line, genes that are required for cell growth and proliferation predominate among the 65% that are TAF7-dependent.Citation19 In resting peripheral T cells, which do not require TAF7 for survival, only a small number (~2.5%) of all transcripts are affected by TAF7 deletion. Of the TAF7-dependent transcripts in mature T cells, many encode components of the TCR signaling pathway.Citation26 Therefore, whereas TAF7 deletion in immature, proliferating cells (i.e., MEFs and DN thymocytes) has a global effect on transcription, its deletion in terminally differentiated, non-proliferating T cells primarily affects a subset of genes related to T cell function. Thus, the cellular effects of TAF7 reflect its transcriptional regulation of the genes required for cell growth and proliferation.
TAF7 is a traffic controller of transcription initiation
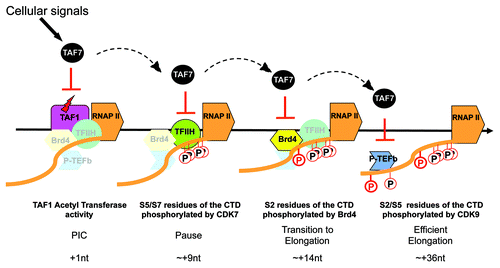
Our studies have revealed a critical role for TAF7 in proliferation and in transcription initiation. Its multiple interactions with components of the RNAP II transcription machinery –TFIID/TAF1, CIITA, BRD4, CDK7/TFIIH, CDK9/PTEFb – all result in the inhibition of their enzymatic activities. Based on our findings, we propose a model in which TAF7 functions as a “traffic controller” in transcription initiation, ensuring that each general transcription factor arrives and functions in the correct sequence (). As we have demonstrated in the case of TAF1, initiation of transcription is accompanied by the release of TAF7 and the recovery of the essential TAF1 AT activity. The inhibition of TAF1 AT by TAF7 during the assembly of the preinitiation complex may serve to prevent premature transcription initiation before all of the necessary factors have been recruited, which would result in abortive termination. We further speculate that the binding of TAF7 and its inhibition of the activity of each of the remaining factors—CDK7, BRD4 and CDK9— plays a similar role, namely to prevent their functioning prematurely. According to this model, TAF7 would be released in a sequential fashion: from TFIIH to enable phosphorylation of the CTD Ser5/7 and the recruitment of capping enzymes, then from BRD4 to allow phosphorylation of the CTD Ser2 and recruitment of splicing factors, and, finally, from PTEFb for the phosphorylation of Ser5/2 and the ongoing recruitment of transcript maturation factors during elongation. In this way, TAF7 regulation of the enzymatic activities of the general transcription factors ensures an orderly progression through both transcription initiation and cell cycle.
Figure 1. TAF7 is a traffic controller of transcription initiation. Schematic representation of TAF7 functions in the early steps of transcription. Before initiation of transcription, TAF7 binds TAF1 and inhibits its AT activity (red lightning bolt) which prevents premature initiation. TAF7 then sequentially regulates TFIIH, Brd4 and p-TEFb, inhibiting each of their kinases activities which blocks CTD phosphorylation and progression of the polymerase. The release of TAF7 from TAF1 and from the CTD kinases allows the RNAP II machinery to move forward. The events inhibited by TAF7, the transcription steps and the positions of the polymerase where they occur are indicated below the schema.
The precise mechanism by which TAF7 exerts its regulatory activity remains to be determined. It is not known if a single molecule of TAF7 is recruited to a preinitiation complex through TFIID and regulates all of the downstream enzymatic activities. Alternatively, each of the CTD kinases could be targeted by separate TAF7 molecules during the assembly of the preinitiation complex and released sequentially. This latter possibility would require a molar excess of TAF7, relative to other TFIID components. Preliminary findings indicate that this is the case: In a variety of cells, there is a molar excess of TAF7 relative to TBP (unpublished observations). In addition, TAF7 has been detected in TFIID free cellular fractions.Citation22
Implicit in the current model is that the different regulatory roles of TAF7 occur on the same gene at different steps of transcription. However, as yet there have been no studies to determine whether all TAF7 functions occur sequentially on a single gene or if TAF7 inhibition of TAF1 AT activity is separable from its inhibition of CTD kinase activities and functioning on different genes.
Future experiments will focus on understanding the precise mechanisms of TAF7 regulation of transcription.
Conclusion
Although originally characterized as a structural component of TFIID, TAF7 is emerging as a critical regulator of transcription initiation and cell proliferation. The molecular mechanisms by which TAF7 blocks proliferation and its direct gene targets in both undifferentiated and differentiated cells remain to be determined. It is interesting to note that the multiplicity of functions already attributed to TAF7 bear a striking parallel to the HIV transactivator Tat. Thus, it is tempting to speculate that TAF7 is the normal cellular homolog of Tat and may, like Tat, have additional functions that remain to be defined.
Acknowledgments
We thank members of our lab for thoughtful discussions and apologize to the researchers whose work could not be referenced due to space limitations. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
References
- Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol 2006; 41:105 - 78; http://dx.doi.org/10.1080/10409230600648736; PMID: 16858867
- Conaway RC, Conaway JW. Function and regulation of the Mediator complex. Curr Opin Genet Dev 2011; 21:225 - 30; http://dx.doi.org/10.1016/j.gde.2011.01.013; PMID: 21330129
- Taatjes DJ. The human Mediator complex: a versatile, genome-wide regulator of transcription. Trends Biochem Sci 2010; 35:315 - 22; http://dx.doi.org/10.1016/j.tibs.2010.02.004; PMID: 20299225
- Dey A, Nishiyama A, Karpova T, McNally J, Ozato K. Brd4 marks select genes on mitotic chromatin and directs postmitotic transcription. Mol Biol Cell 2009; 20:4899 - 909; http://dx.doi.org/10.1091/mbc.E09-05-0380; PMID: 19812244
- Devaiah BN, Lewis BA, Cherman N, Hewitt MC, Albrecht BK, Robey PG, et al. BRD4 is an atypical kinase that phosphorylates serine2 of the RNA polymerase II carboxy-terminal domain. Proc Natl Acad Sci U S A 2012; 109:6927 - 32; http://dx.doi.org/10.1073/pnas.1120422109; PMID: 22509028
- Egloff S, Dienstbier M, Murphy S. Updating the RNA polymerase CTD code: adding gene-specific layers. Trends Genet 2012; 28:333 - 41; http://dx.doi.org/10.1016/j.tig.2012.03.007; PMID: 22622228
- Hsin JP, Manley JL. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev 2012; 26:2119 - 37; http://dx.doi.org/10.1101/gad.200303.112; PMID: 23028141
- Cler E, Papai G, Schultz P, Davidson I. Recent advances in understanding the structure and function of general transcription factor TFIID. Cell Mol Life Sci 2009; 66:2123 - 34; http://dx.doi.org/10.1007/s00018-009-0009-3; PMID: 19308322
- D’Alessio JA, Wright KJ, Tjian R. Shifting players and paradigms in cell-specific transcription. Mol Cell 2009; 36:924 - 31; http://dx.doi.org/10.1016/j.molcel.2009.12.011; PMID: 20064459
- Müller F, Zaucker A, Tora L. Developmental regulation of transcription initiation: more than just changing the actors. Curr Opin Genet Dev 2010; 20:533 - 40; http://dx.doi.org/10.1016/j.gde.2010.06.004; PMID: 20598874
- Dikstein R, Ruppert S, Tjian R. TAFII250 is a bipartite protein kinase that phosphorylates the base transcription factor RAP74. Cell 1996; 84:781 - 90; http://dx.doi.org/10.1016/S0092-8674(00)81055-7; PMID: 8625415
- Mizzen CA, Yang XJ, Kokubo T, Brownell JE, Bannister AJ, Owen-Hughes T, et al. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell 1996; 87:1261 - 70; http://dx.doi.org/10.1016/S0092-8674(00)81821-8; PMID: 8980232
- Gegonne A, Weissman JD, Zhou M, Brady JN, Singer DS. TAF7: a possible transcription initiation check-point regulator. Proc Natl Acad Sci U S A 2006; 103:602 - 7; http://dx.doi.org/10.1073/pnas.0510031103; PMID: 16407123
- Dunphy EL, Johnson T, Auerbach SS, Wang EH. Requirement for TAF(II)250 acetyltransferase activity in cell cycle progression. Mol Cell Biol 2000; 20:1134 - 9; http://dx.doi.org/10.1128/MCB.20.4.1134-1139.2000; PMID: 10648598
- Hilton TL, Li Y, Dunphy EL, Wang EH. TAF1 histone acetyltransferase activity in Sp1 activation of the cyclin D1 promoter. Mol Cell Biol 2005; 25:4321 - 32; http://dx.doi.org/10.1128/MCB.25.10.4321-4332.2005; PMID: 15870300
- Weissman JD, Howcroft TK, Singer DS. TAF(II)250-independent transcription can be conferred on a TAF(II)250-dependent basal promoter by upstream activators. J Biol Chem 2000; 275:10160 - 7; http://dx.doi.org/10.1074/jbc.275.14.10160; PMID: 10744699
- Gegonne A, Weissman JD, Singer DS. TAFII55 binding to TAFII250 inhibits its acetyltransferase activity. Proc Natl Acad Sci U S A 2001; 98:12432 - 7; http://dx.doi.org/10.1073/pnas.211444798; PMID: 11592977
- Kloet SL, Whiting JL, Gafken P, Ranish J, Wang EH. Phosphorylation-dependent regulation of cyclin D1 and cyclin A gene transcription by TFIID subunits TAF1 and TAF7. Mol Cell Biol 2012; 32:3358 - 69; http://dx.doi.org/10.1128/MCB.00416-12; PMID: 22711989
- Devaiah BN, Lu H, Gegonne A, Sercan Z, Zhang H, Clifford RJ, et al. Novel functions for TAF7, a regulator of TAF1-independent transcription. J Biol Chem 2010; 285:38772 - 80; http://dx.doi.org/10.1074/jbc.M110.173864; PMID: 20937824
- Shen WC, Bhaumik SR, Causton HC, Simon I, Zhu X, Jennings EG, et al. Systematic analysis of essential yeast TAFs in genome-wide transcription and preinitiation complex assembly. EMBO J 2003; 22:3395 - 402; http://dx.doi.org/10.1093/emboj/cdg336; PMID: 12840001
- Raval A, Howcroft TK, Weissman JD, Kirshner S, Zhu XS, Yokoyama K, et al. Transcriptional coactivator, CIITA, is an acetyltransferase that bypasses a promoter requirement for TAF(II)250. Mol Cell 2001; 7:105 - 15; http://dx.doi.org/10.1016/S1097-2765(01)00159-9; PMID: 11172716
- Gegonne A, Weissman JD, Lu H, Zhou M, Dasgupta A, Ribble R, et al. TFIID component TAF7 functionally interacts with both TFIIH and P-TEFb. Proc Natl Acad Sci U S A 2008; 105:5367 - 72; http://dx.doi.org/10.1073/pnas.0801637105; PMID: 18391197
- Yamada T, Yamaguchi Y, Inukai N, Okamoto S, Mura T, Handa H. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol Cell 2006; 21:227 - 37; http://dx.doi.org/10.1016/j.molcel.2005.11.024; PMID: 16427012
- Devaiah BN, Singer DS. Cross-talk among RNA Polymerase II kinases modulates C-terminal domain phosphorylation. J Biol Chem 2012; 287:38755 - 66; http://dx.doi.org/10.1074/jbc.M112.412015; PMID: 23027873
- Bosselut R. CD4/CD8-lineage differentiation in the thymus: from nuclear effectors to membrane signals. Nat Rev Immunol 2004; 4:529 - 40; http://dx.doi.org/10.1038/nri1392; PMID: 15229472
- Gegonne A, Tai X, Zhang J, Wu G, Zhu J, Yoshimoto A, et al. The general transcription factor TAF7 is essential for embryonic development but not essential for the survival or differentiation of mature T cells. Mol Cell Biol 2012; 32:1984 - 97; http://dx.doi.org/10.1128/MCB.06305-11; PMID: 22411629
- Hayashida T, Sekiguchi T, Noguchi E, Sunamoto H, Ohba T, Nishimoto T. The CCG1/TAFII250 gene is mutated in thermosensitive G1 mutants of the BHK21 cell line derived from golden hamster. Gene 1994; 141:267 - 70; http://dx.doi.org/10.1016/0378-1119(94)90583-5; PMID: 8163200
- Mochizuki K, Nishiyama A, Jang MK, Dey A, Ghosh A, Tamura T, et al. The bromodomain protein Brd4 stimulates G1 gene transcription and promotes progression to S phase. J Biol Chem 2008; 283:9040 - 8; http://dx.doi.org/10.1074/jbc.M707603200; PMID: 18223296