Abstract
Mesp1 sits on the tip of the cardiac regulatory hierarchy, recent evidences support that it is regulated by stem cell factor Oct4, early gastrulation signal canonical Wnts and a couple of T-box factors, T and Eomes. With other transcription factors, Mesp1 programs/reprograms human cells toward cardiomyocytes.
Lessons learned from cardiopoiesis has directed our attention to the serial programming that controls the appearances of cardiac progenitors from embryonic stem cells. Aggregation of ES cells into hanging drop embryoid bodies is an initial step to inhibit stem cell activity and generate endodermal cell signaling with the activation of Sox 17 and Hhex and the early appearance of the mesodermal markers T-brachyury and goosecoid. Subsequently transient induction of Mesp1 and Mesp2, a pair of related basic helix–loop–helix transcription factors, is indispensable for creating heart mesoderm. Next, the appearances of core cardiac transcription factors led by Nkx2.5, Isl1, Gata4, Mef2C and Tbx5 convert these embryonic cells into cardiac progenitors usually followed by the appearance of contractile structural genes, sarcomere formation and rhythmic beating. In early vertebrate embryos, deletion mutagenesis of these core TFs, however, often leads to cardiac structural defects (cardia bifida, looping defects, etc.) instead of the lack of heart formation or hears in other anatomical sites, which is best explained by redundant, parallel and compensatory roles of these TFs. Strikingly, Gata4, Mef2c and Tbx5 were shown to reprogram mouse fibroblast into cardiomyocytes, both in cell culture and in post-myocardium infarction hearts.Citation1-Citation3 Recently, the idea to reprogram fibroblasts by using fundamental regulatory paradigms has engendered a great deal of interest. Trans-differentiation, the conversion of cell phenotypes by selective enhancement of gene expression, is a new path that may control the terminal commitment of cells.Citation4,Citation5 Whether the same sets of TFs will reprogram human cells is to be determined, but this discovery resoundingly testified the critical position of cardiac TFs in reprograming and cardiac therapy.
Sitting on the tip of the cardiac TF hierarchy is the bHLH factor Mesp1. It is first expressed at the onset of gastrulation, along the primitive streak and in the premesoderm that eventually gives rise to the heart. Lineage tracing studies using Cre recombinase knocked into the Mesp1 locus revealed that Mesp1-expressing cells mainly contribute to the mesoderm component of the amnion, and to the myocardium of the heart tube.Citation6,Citation7 Homologous disruption of the Mesp1 gene resulted in aberrant cardiac morphogenesis. Mesp1-expressing cells are delayed in migrating from the primitive streak to the heart field, in the homozygous Mesp1-deficient embryos.Citation6,Citation7 Furthermore, simultaneous disruption of both Mesp1 and Mesp2 genes (dKO) led to complete loss of posterior structures including heart, somites and gut. Chimera analysis, however, showed that Mesp1 and Mesp2 dKO cells still contributed to the formation of somites and gut, but not heart.Citation8 Mesp1 drives mouse ES cells toward the cardiac fate. Transient expression of Mesp1 accelerated and enhanced the appearance of cardiac progenitors.Citation9-Citation11 Several transcription factors with known cardiac function, including Hand2, Gata4, Myocardin, FoxH1, FoxC1 were upregulated within 18 h following Mesp1 induction. To date, Mesp1 is recognized as the first sign of the cardiac lineages out of the nascent mesoderm.
Despite the pivotal role of Mesp1 in cardiogenesis, its expression is only transient in the developing embryo. Mesp1 starts to express at the onset of mesoderm formation (E6.0) and almost completely disappears before the appearance of cardiac crescent (E7.0), which suggests that Mesp1 is not required for later stages of cardiac development.Citation6,Citation7 So, how is Mesp1 gene expression regulated? Only recently two enhancer regions on the promoter of Mesp1 were identified to drive Mesp1 expression in the early mesoderm. Elucidation of the regulatory trans-acting transcription factors is being pursued.Citation12,Citation13 Whether Mesp1 is capable to reprogram non-cardiac cells toward the cardiac fate remains obscure, though Mesp1 seemed to enhance the reprograming efficiency of Gata4, Mef2c and Tbx5.Citation1
Mesp1 is directly regulated by early developmental signals
By analyzing a 6012 bp upstream region of the Mesp1 gene, we have identified a putative Tcf/Lef-Oct4 composite site (-31 to -17 relative to transcription starting site)().Citation14 The putative Tcf/Lef site is a perfect match to the Tcf/Lef consensus sequence CTTTG[AT][AT].Citation15 And it sits adjacent to an “octamer” putative Oct4 binding site. Such a composite site is similar to the previously identified Sox2-Oct4 composite site, residing on the promoters of Sox2, Oct4, Nanog and a few other genes related to pluripotency.Citation16 In reporter assays, Oct4 only marginally transactivated the reporter, but had a potent synergistic effect with CA-β-Cat. Consistently, γ-32P-labeled oligos harboring the Tcf/Lef-Oct4 element and in vitro translated Oct4 and Lef1 proteins formed a ternary complex in gel shift assays. The specificity of binding was confirmed by excess amount of unlabeled oligos (wild type, mutants affect the Tcf/Lef site, the Oct4 site or both) and specific antibodies.
Figure 1. A Tcf/Lef-Oct4 site mediates the cooperation of Oct4 and canonical Wnt on the Mesp1 promoter. (A) Schematic diagram of conserved transcription factor binding site predicted by rVista. The Tcf/Lef-Oct4 site and adjacent sequence are shown. Blue characters mark the Tcf/Lef site, and red mark the “octamer” Oct4 site. Shaded characters mark the point mutations used in this study. (B) Constitutively active β-Catenin stimulated the Mesp1-Luc reporter in a dose-dependent manner. (C) Oct4 synergized with CA-β-Cat in co-activating the Mesp1-Luc reporter, albeit Oct4 only marginally activated it. (D) Mutations on the Tcf/Lef-Oct4 site impaired Mesp1-LacZ expression in a transient transgenic assay. The table summarizes the number of β-galactosidase positives vs. genotype positives in each wildtype or mutant group. Images on the bottom show typical results of the LacZ staining from each group. (E) A working model of Oct4’s role in sustaining pluripotency and inducing differentiation. Through sequence-identical composite sites, a Sox2-Oct4 tandem and a Tcf/Lef-Oct4 tandem regulate pluripotency and differentiation, respectively.
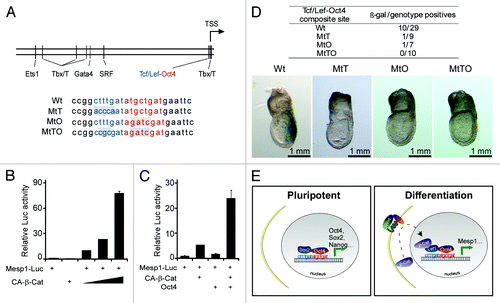
Transient transgenic assays have revealed critical function of the Tcf/Lef-Oct4 site on Mesp1 expression during embryogenesis. The 6012 bp promoter of Mesp1 was fused to a LacZ reporter gene, and this construct was injected into the pronuclei of fertilized eggs. The resulted E7.5 embryos were analyzed for β-galactosidase expression. The wild type promoter led to β-galactosidase expression in the primitive streak, where Mesp1 mRNA was shown to be expressed at peak levels.Citation7 Mutation on both Tcf/Lef and “octamer” side of the composite site resulted in complete loss of β-galactosidase staining.
From these findings we have learned that Oct4 plays an active role in transactivating Mesp1, by integrating a crucial gastrulation signal in Wnt, which also provides a reasonable explanation for the observations that elevated Oct4 triggers cardiac differentiation.Citation17-Citation20 This Oct4/Wnt signaling model represents an extra dimension of regulation over the switch from pluripotency to differentiation. It ensures that the activation of lineage-related genes such as Mesp1 is only possible in progenitor cells, where a burst of canonical Wnt signaling meets with residual Oct4 expression. In other cell types, for example, terminally differentiated cells, the ability to activate key lineage genes like Mesp1 is compromised due to the loss of Oct4 expression.
As pointed out earlier, the Tcf/Lef-Oct4 composite element is similar to a Sox2-Oct4 composite element found in a few pluripotent genes. Furthermore, Tcf/Lef and Sox are most closely related within the High-Mobility Group (HMG) protein family. They use a single HMG domain to bind to the minor groove of DNA and cause it to bend.Citation21 This prompts an intriguing notion that a Tcf/Lef-Sox2 swap may switch off pluripotency and switch on differentiation. We proposed a working model of Oct4 in regulating pluripotency and differentiation. In a number of pluripotent genes (Oct4, Sox2, Nanog, and others), Oct4 and Sox2 may co-occupy a composite site and activate transcription of these genes to maintain pluripotency. In lineage-related genes such as Mesp1, Oct4 and Tcf/Lef co-occupy a similar composite site. During differentiation, a burst of canonical Wnt signaling leads to the accumulation of β-Catenin in the nucleus, which results in synergistic activation of the lineage-related genes ().
Consistent with the temporal appearance of Mesp1 during embryogenesis, Mesp1 has been shown to be regulated by gastrulation factors T and Eomes, through multiple T-box elements identified on the Mesp1 promoter. Mesp1+ cells were shown to emerge from a T+ population in which shRNA targeting of T greatly impaired Mesp1 expression.Citation22 Comparably, Eomes also directly targets T-box elements on the Mesp1 promoter, and acts upstream of Mesp1 to specify the cardiac mesoderm.Citation23,Citation24 However, Eomes’s role in cardiac differentiation is contingent upon the level of Activin/Nodal. High levels of Activin inhibit cardiac mesoderm formation by Eomes, and favor Eomes-mediated definitive endoderm formation.Citation24
These recent developments in our understanding of the early cardiac transcription events fill in a previously unexplored area. They continue to pave the way leading to using molecular cues to direct the cardiac cell fate. These studies still have not answered a key question: Given the relatively broad expression of stem cell factors and early gastrulation factors, how do these factors specify the Mesp1-lineage of cardiac progenitor cells?
Mesp1 is a core factor in programing and reprograming toward the cardiac fate in human cells
At the front of cardiac reprograming, Gata4, Mef2c and Tbx5 were shown to convert murine fibroblasts to cardiomyocytes.Citation2,Citation3 Addition of Hand2 to the mix increased the rate of conversion in an independent study.Citation1 These results are surprising as two TFs that have perhaps more defined role in the primary and secondary heart field, Nkx2–5 and Isl1, are out of the loop. Addition of Mesp1 seemed to have little effect in increasing the conversion rate. It is worth noting that the factors of choice depend largely on the screening strategy and the original cell type to be reprogramed. For example, it was reported that Myocardin, in addition to Mef2c and Tbx5 was most effective in cardiac reprograming, but Gata4 in addition to MT was detrimental, which was contradictory to an earlier report.Citation25 The lack of success of GMT thus far in converting human cells suggests that the program of human somatic cells are of higher complexity than rodent cells.
Given the undisputed importance of Mesp1 in early cardiac development, we had attempted to use Mesp1 to reprogram human dermal fibroblasts. Use of Mesp1 alone had not yielded significant changes in cell morphology or cardiac gene expression. Since Mesp1 is also conserved in chordates, we resourced to Ciona Intestinalis for additional factors that are well conserved within chordates (Ciona and vertebrates) and co-operate with Mesp1 in cardiac fate decisions. There is a single Mesp ortholog, Ci-Mesp, in Ciona. It is strictly expressed in the pre-cardiac lineages at the 110-cell stage. Shortly after Ci-Mesp1, Ets1/2 is expressed, which defines the pre-cardiac mesoderm and makes the cells responsive to heart-specifying signals.Citation26 We thus explored the combination of Mesp1/Ets2 in reprogramming human cells. Although neither ETS2 nor MESP1 can by themselves generate cardiac progenitors de novo from fibroblasts, forced co-expression of ETS2 and MESP1 or treatment with purified ETS2 and MESP1 proteins reprograms fibroblasts into cardiac progenitors, as shown by the de novo appearance of core cardiac transcription factors, Ca2+ transients, and sarcomeres ().Citation27
Figure 2. MESP1 and ETS2 reprogram human dermal fibroblasts into cardiac progenitor cells. (A) A reprogramming protocol using purified TAT-ETS2 and TAT-MESP1 to transduce NHDFs. (B) Within 8 d after the beginning of combined ETS2 and MESP1 protein treatment, NKX2.5-tdTomato+ cells appeared in colonies of cellular aggregates. Magnification, 10 × . (C) FACS analysis revealed a conversion of about 9% of the NHDFs that were tdTomato+. (D) Percentages of spontaneously beating cells after Dox-induced conversion of NHDFs by ETS2 and MESP1 in the absence (O) and presence of Activin A (A) and BMP2 (B). (E) Model for reprogramming of human fibroblasts into cardiac progenitors induced by overexpression of ETS2 and MESP1 or treatment with purified proteins, as supported by data from our study.
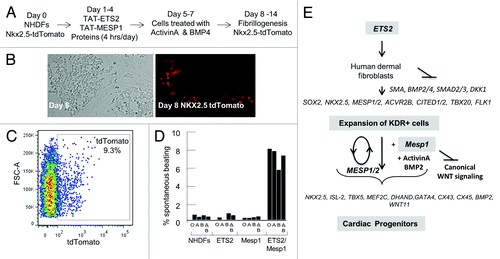
Compared with Ciona Ets1/2, human ETS2 seems to have additional roles, which unexpectedly favor cardiac reprograming. Constitutive ETS2 overexpression in NHDFs gave rise to highly replicative small rounded cells within one week after lentiviral infection. ETS2-induced the appearance of cell types with some of the characteristics of endothelial and cardiac progenitors. They express surface markers CD31/platelet endothelial cell adhesion molecule 1 (PECAM1) and CD309/kinase insert domain receptor (KDR). Furthermore, ETS2 was able to enrich the RNA transcripts of core components in the Activin/Nodal program (ACVR2B, TDGF1, CITED2, and LEFTY1). The Activin/Nodal pathway is obligatory for the appearance of cardiac progenitors and in its absence cell fate is shifted toward a neural program. Expression of the core cardiac transcription factors, including NKX2.5, MESP1, MEF2C, GATA4, and ISL1 was also stimulated by ETS2. However, these KDR+ cells did not express smooth muscle actin (SMA/ACTA2), SM22α/TGLN, or induce terminal cardiogenesis. Only with co-expressed MESP1, or MESP1 protein, these cells showed the classic characters of cardiomyocytes.
When the embryonic mouse heart develops from a “primitive heart tube” at E8.5 to a well-defined tubular shape expanded at the ventricular end at E9, Ca2+ activity increases from 50% to almost 100% of the nascent myocytesCitation28 and electrophysiological maturation of human ES-derived cardiomyocytes involves the development of more rapid spontaneous Ca2+ transients.Citation29 In comparison, the ETS2/MESP1 reprogrammed cells appear to be at the early to intermediate stages of cardiomyocyte maturation. The ETS2/MESP1-transdifferentiated cells can be considered cardiac progenitors at a baseline that may need to be further maturated into terminal differentiated cardiomyocytes, likely by using co-cultures with cardiomyocytes and stimulation with protein factors and small molecules.Citation30,Citation31
In human ES and iPS cells, Mesp1 was shown to require other early cardiac factors for forward programing the cells to cardiomyocytes. Focused screening of TF combinations revealed that BAF60C, GATA4, and MESP1 (BGM) were most effective for cardiac forward programming in human induced pluripotent stem cell lines and human ES cells. Each of these TFs alone had no inductive effect. The combination of GATA4 and MESP1 was required for the forward programing, whereas BAF60C depletion only slightly diminished formation of CM-like cells.Citation32
With only limited number of cardiac programing reports, it is hard to determine if the current reprograming cocktails represent the most effective, especially for comparison between species. The two examples here suggest that Mesp1, despite ineffectiveness when used alone, is a key factor in programing/reprogramming human cells. Further development upon the current regimen may require the answering of several inter-related key questions. First, what are other factors that co-operate with Mesp1 in determining the early cardiac cell fate? The human ETS family comprises 29 members, among which ETS1, FLI1, ETV1, ETV5, ERG, and ETV6 are the most abundant in the early embryonic heart, and perhaps are better MESP1 partners.Citation33 GATA4 is a TF with functions in the endoderm as well as in the cardiac lineages.Citation34 The concurrent requirement of both GATA4 and MESP1 for forward programing in human ES and iPS cells may be alternatively explained by the need for de novo cardiac mesoderm specification and cardiogenic endoderm signals. Finding an optimized reprograming for MESP1 may ultimately depend on the understanding of the earliest factors in the de novo induction cardiac mesoderm events, such as those described in the first half of this article. Second, whether MESP1 is continuously required for cardiac programing/reprograming. The early disappearance of MESP1 suggests that it is not required for later stage of cardiac development. In Ciona, the role of MESP1 in cardiac mesoderm specification and mesoderm migration are separated.Citation35 Answering this question is directly connected to the timing of MESP1 in programing/reprograming procedures. Finally, how are downstream lineages of MEPS1-expressing mesoderm separated? Our preliminary results suggest that Mesp1-expressing mesoderm differentiates into other cell lineages in addition to the cardiovascular system. Dissecting the molecular pathways that regulate the separation of downstream lineages would allow generation of the optimized cell types for future therapeutic studies.
| Abbreviations: | ||
| TFs | = | Transcription Factors |
| bHLH | = | basic Helix-Loop-Helix |
| dKO | = | double Knock Out |
| ES | = | Embryonic Stem |
| CA-β-Cat | = | Constitutively Active- β-Catenin |
| HMG | = | High Mobility Group |
| CM | = | Cardiomyocytes |
| shRNA | = | small hairpin RNA |
Acknowledgments
Research from our lab discussed in this article is supported by research funds from NIH (RJS), the Texas Heart Institute (RJS), the University of Houston (RJS) and from the American Heart Association (YL).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Song K, Nam YJ, Luo X, Qi X, Tan W, Huang GN, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 2012; 485:599 - 604; http://dx.doi.org/10.1038/nature11139; PMID: 22660318
- Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 2012; 485:593 - 8; http://dx.doi.org/10.1038/nature11044; PMID: 22522929
- Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 2010; 142:375 - 86; http://dx.doi.org/10.1016/j.cell.2010.07.002; PMID: 20691899
- Baeyens L, De Breuck S, Lardon J, Mfopou JK, Rooman I, Bouwens L. In vitro generation of insulin-producing beta cells from adult exocrine pancreatic cells. Diabetologia 2005; 48:49 - 57; http://dx.doi.org/10.1007/s00125-004-1606-1; PMID: 15616797
- Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature 2008; 455:627 - 32; http://dx.doi.org/10.1038/nature07314; PMID: 18754011
- Saga Y, Kitajima S, Miyagawa-Tomita S. Mesp1 expression is the earliest sign of cardiovascular development. Trends Cardiovasc Med 2000; 10:345 - 52; http://dx.doi.org/10.1016/S1050-1738(01)00069-X; PMID: 11369261
- Saga Y, Miyagawa-Tomita S, Takagi A, Kitajima S, Miyazaki Ji, Inoue T. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development 1999; 126:3437 - 47; PMID: 10393122
- Kitajima S, Takagi A, Inoue T, Saga Y. MesP1 and MesP2 are essential for the development of cardiac mesoderm. Development 2000; 127:3215 - 26; PMID: 10887078
- David R, Brenner C, Stieber J, Schwarz F, Brunner S, Vollmer M, et al. MesP1 drives vertebrate cardiovascular differentiation through Dkk-1-mediated blockade of Wnt-signalling. Nat Cell Biol 2008; 10:338 - 45; http://dx.doi.org/10.1038/ncb1696; PMID: 18297060
- Lindsley RC, Gill JG, Murphy TL, Langer EM, Cai M, Mashayekhi M, et al. Mesp1 coordinately regulates cardiovascular fate restriction and epithelial-mesenchymal transition in differentiating ESCs. Cell Stem Cell 2008; 3:55 - 68; http://dx.doi.org/10.1016/j.stem.2008.04.004; PMID: 18593559
- Bondue A, Lapouge G, Paulissen C, Semeraro C, Iacovino M, Kyba M, et al. Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell 2008; 3:69 - 84; http://dx.doi.org/10.1016/j.stem.2008.06.009; PMID: 18593560
- Haraguchi S, Kitajima S, Takagi A, Takeda H, Inoue T, Saga Y. Transcriptional regulation of Mesp1 and Mesp2 genes: differential usage of enhancers during development. Mech Dev 2001; 108:59 - 69; http://dx.doi.org/10.1016/S0925-4773(01)00478-6; PMID: 11578861
- Oginuma M, Hirata T, Saga Y. Identification of presomitic mesoderm (PSM)-specific Mesp1 enhancer and generation of a PSM-specific Mesp1/Mesp2-null mouse using BAC-based rescue technology. Mech Dev 2008; 125:432 - 40; http://dx.doi.org/10.1016/j.mod.2008.01.010; PMID: 18328678
- Li Y, Yu W, Cooney AJ, Schwartz RJ, Liu Y. Oct4 and Canonical Wnt Signaling Regulate the Cardiac Lineage Factor Mesp1 through a Tcf/Lef-Oct4 Composite Element. Stem Cells 2013; http://dx.doi.org/10.1002/stem.1362; PMID: 23417899
- Grosschedl R, Giese K, Pagel J. HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet 1994; 10:94 - 100; http://dx.doi.org/10.1016/0168-9525(94)90232-1; PMID: 8178371
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 2008; 133:1106 - 17; http://dx.doi.org/10.1016/j.cell.2008.04.043; PMID: 18555785
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet 2000; 24:372 - 6; http://dx.doi.org/10.1038/74199; PMID: 10742100
- Blin G, Nury D, Stefanovic S, Neri T, Guillevic O, Brinon B, et al. A purified population of multipotent cardiovascular progenitors derived from primate pluripotent stem cells engrafts in postmyocardial infarcted nonhuman primates. J Clin Invest 2010; 120:1125 - 39; http://dx.doi.org/10.1172/JCI40120; PMID: 20335662
- Stefanovic S, Abboud N, Désilets S, Nury D, Cowan C, Pucéat M. Interplay of Oct4 with Sox2 and Sox17: a molecular switch from stem cell pluripotency to specifying a cardiac fate. J Cell Biol 2009; 186:665 - 73; http://dx.doi.org/10.1083/jcb.200901040; PMID: 19736317
- Zeineddine D, Papadimou E, Chebli K, Gineste M, Liu J, Grey C, et al. Oct-3/4 dose dependently regulates specification of embryonic stem cells toward a cardiac lineage and early heart development. Dev Cell 2006; 11:535 - 46; http://dx.doi.org/10.1016/j.devcel.2006.07.013; PMID: 17011492
- Laudet V, Stehelin D, Clevers H. Ancestry and diversity of the HMG box superfamily. Nucleic Acids Res 1993; 21:2493 - 501; http://dx.doi.org/10.1093/nar/21.10.2493; PMID: 8506143
- David R, Jarsch VB, Schwarz F, Nathan P, Gegg M, Lickert H, et al. Induction of MesP1 by Brachyury(T) generates the common multipotent cardiovascular stem cell. Cardiovasc Res 2011; 92:115 - 22; http://dx.doi.org/10.1093/cvr/cvr158; PMID: 21632880
- Costello I, Pimeisl IM, Dräger S, Bikoff EK, Robertson EJ, Arnold SJ. The T-box transcription factor Eomesodermin acts upstream of Mesp1 to specify cardiac mesoderm during mouse gastrulation. Nat Cell Biol 2011; 13:1084 - 91; http://dx.doi.org/10.1038/ncb2304; PMID: 21822279
- van den Ameele J, Tiberi L, Bondue A, Paulissen C, Herpoel A, Iacovino M, et al. Eomesodermin induces Mesp1 expression and cardiac differentiation from embryonic stem cells in the absence of Activin. EMBO Rep 2012; 13:355 - 62; http://dx.doi.org/10.1038/embor.2012.23; PMID: 22402664
- Protze S, Khattak S, Poulet C, Lindemann D, Tanaka EM, Ravens U. A new approach to transcription factor screening for reprogramming of fibroblasts to cardiomyocyte-like cells. J Mol Cell Cardiol 2012; 53:323 - 32; http://dx.doi.org/10.1016/j.yjmcc.2012.04.010; PMID: 22575762
- Davidson B, Shi W, Beh J, Christiaen L, Levine M. FGF signaling delineates the cardiac progenitor field in the simple chordate, Ciona intestinalis. Genes Dev 2006; 20:2728 - 38; http://dx.doi.org/10.1101/gad.1467706; PMID: 17015434
- Islas JF, Liu Y, Weng KC, Robertson MJ, Zhang S, Prejusa A, et al. Transcription factors ETS2 and MESP1 transdifferentiate human dermal fibroblasts into cardiac progenitors. Proc Natl Acad Sci U S A 2012; 109:13016 - 21; http://dx.doi.org/10.1073/pnas.1120299109; PMID: 22826236
- Chen F, De Diego C, Chang MG, McHarg JL, John S, Klitzner TS, et al. Atrioventricular conduction and arrhythmias at the initiation of beating in embryonic mouse hearts. Dev Dyn 2010; 239:1941 - 9; http://dx.doi.org/10.1002/dvdy.22319; PMID: 20549739
- Kim C, Majdi M, Xia P, Wei KA, Talantova M, Spiering S, et al. Non-cardiomyocytes influence the electrophysiological maturation of human embryonic stem cell-derived cardiomyocytes during differentiation. Stem Cells Dev 2010; 19:783 - 95; http://dx.doi.org/10.1089/scd.2009.0349; PMID: 20001453
- Willems E, Lanier M, Forte E, Lo F, Cashman J, Mercola M. A chemical biology approach to myocardial regeneration. J Cardiovasc Transl Res 2011; 4:340 - 50; http://dx.doi.org/10.1007/s12265-011-9270-6; PMID: 21424858
- Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, et al. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell 2011; 8:228 - 40; http://dx.doi.org/10.1016/j.stem.2010.12.008; PMID: 21295278
- Hartung S, Schwanke K, Haase A, David R, Franz WM, Martin U, et al. Directing cardiomyogenic differentiation of human pluripotent stem cells by plasmid-based transient overexpression of cardiac transcription factors. Stem Cells Dev 2013; 22:1112 - 25; http://dx.doi.org/10.1089/scd.2012.0351; PMID: 23157212
- Hollenhorst PC, McIntosh LP, Graves BJ. Genomic and biochemical insights into the specificity of ETS transcription factors. Annu Rev Biochem 2011; 80:437 - 71; http://dx.doi.org/10.1146/annurev.biochem.79.081507.103945; PMID: 21548782
- Holtzinger A, Rosenfeld GE, Evans T. Gata4 directs development of cardiac-inducing endoderm from ES cells. Dev Biol 2010; 337:63 - 73; http://dx.doi.org/10.1016/j.ydbio.2009.10.003; PMID: 19850025
- Davidson B, Shi W, Levine M. Uncoupling heart cell specification and migration in the simple chordate Ciona intestinalis. Development 2005; 132:4811 - 8; http://dx.doi.org/10.1242/dev.02051; PMID: 16207759