Abstract
New approaches using biotinylated-psoralen as a probe for investigating DNA structure have revealed new insights into the relationship between DNA supercoiling, transcription and chromatin compaction. We explore a hypothesis that divergent RNA transcription generates negative supercoiling at promoters facilitating initiation complex formation and subsequent promoter clearance.
Packaging DNA into chromatin creates a repressive environment. This is thought to reduce transcriptional noise and provide additional levels of gene regulation. Many processes have been described that alter chromatin architecture and facilitate or repress transcription. These include transcription factor binding, recruitment of RNA polymerases, ATP-dependent chromatin remodeling and changes in histone modifications. The level of DNA supercoiling is also known to be important for transcription but remains poorly understood, as few techniques are available for directly investigating it. This is about to change: new approaches have recently been used for analyzing DNA topology in cellsCitation1-Citation3 and show transcription dependent changes in DNA supercoiling that impact on higher levels of chromatin structure. Furthermore, new data indicates that DNA supercoiling and divergent RNA transcription are linked providing new ideas for understanding the role of DNA topology in regulating gene transcription.
The structure of DNA within mammalian cells is not known, although it is often thought to be in a B-form configuration. However, this will depend on the base composition, extent of supercoilingCitation4 and DNA binding proteins, such as HMGs. As DNA moves through the RNA polymerase positive (over-wound) supercoils are generated ahead of the polymerase and negative (under-wound) supercoils behind, described as the twin supercoiled domain model.Citation5 Therefore, transcription generates significant torsion in the DNA, so the observation that topoisomerase I, an enzyme that relaxes supercoils, is associated with the transcription machineryCitation6 and localizes to actively transcribed chromatinCitation7 led to the suggestion that it might relieve topological strain. However, early experiments show that genes are more efficiently transcribed if they are encoded on negatively supercoiled plasmids.Citation8,Citation9 In addition, superhelical tension is also a prerequisite for TFIID bindingCitation10 promoting the formation of a preinitiation complex,Citation11 transcription factor bindingCitation4 and promoter clearance.Citation12 Consequently, cell based experiments using psoralen, as a probe for DNA supercoiling, show that promoter regions are under-woundCitation1,Citation13 and supercoiling may prime specific genes for transcription.Citation14 Together this suggests that DNA supercoiling must be precisely controlled and is an additional level of transcriptional regulation. However, how DNA supercoiling is introduced at TSS prior to transcription remains unanswered.
A major clue arose from an in vivo study demonstrating that local domains of supercoiling can affect transcription.Citation15 In this work, a promoter is activated due to transcription generated negative supercoils from a nearby divergent promoter and inhibition of endogenous topoisomerase I further increases transcription. While this study exploited in vitro generated plasmid templates, divergent transcription, whereby transcription is initiated in both sense and anti-sense directions from a promoter, has been shown to be a feature of many active mammalian genes.Citation16-Citation18 The high resolution mapping of short RNA transcripts reveals extensive sense and anti-sense transcript peaks at approximately 50 bp and 250 bp respectively from the TSS. Antisense RNAs are cappedCitation19 but are present at a relatively low abundance as they are substrates for the exosome.Citation18 Histone modifications reflective of transcription initiation (H3K4me3) are found both in the sense and antisense direction while histone marks indicative of transcriptional elongation (H3K79me2) are found in the sense direction.Citation17,Citation20 Consequently divergent transcription is more appropriately described as divergent initiation as only productive elongation occurs in the direction of the gene.
Divergent initiation is very prevalent (approximately 80% of transcribed mammalian genes) but does it have a regulatory function or can it just represent “sloppy” transcription initiation events resulting from an open promoter architecture? Several functions have been proposedCitation21 but, in particular, Seila et al.Citation22 suggest divergent initiation may under-wind promoter regions facilitating productive transcription initiation and elongation. To better investigate DNA supercoiling we have developed an approach for directly analyzing it in cells.Citation1 This technique builds on previous studies that have exploited the cell permeable drug tri-methyl psoralen (TMP), which preferentially intercalates and can be cross-linked into negatively supercoiled (under-wound) DNA.Citation23 The addition of a biotin group, attached via a linker, to the psoralen (bTMP) enables the selective enrichment of under-wound DNA fragments that can be mapped by hybridization to high-resolution genomic microarrays. In RPE1 cells, bTMP binding analysis demonstrates that TSSs are under topological strain and negatively supercoiled in a region that extends ~20 kb into the body of the gene and 10 kb upstream (). Transcription inhibition by α-amanitin or DRBCitation2 causes significant remodeling of TSS DNA to a more positively supercoiled state and subsequent α-amanitin washout promotes a remodeling of the TSS back to its under-wound level. Treatment of cells with bleomycin (introducing DNA nicks) relaxes DNA supercoiling demonstrating that promoter regions are under topological tension, which is also regulated by topoisomerase I and II.Citation1,Citation2
Figure 1. Changes in transcription alter DNA supercoiling. (A) Diagram showing inhibition and recovery of transcription after α-amanitin treatment and meta-analysis showing DNA supercoiling around transcription start sites before, during and after transcription inhibition. For methods see reference Citation1. (B) western blot showing levels of RNA polymerase after α-amanitin treatment in the presence and absence of MG132, a proteasome inhibitor. Cells were suspended in 2 x SDS lysis buffer, incubated at 100°C for 5 min and sonicated briefly. Protein samples were resolved on 8% bis-tris gels and transferred to PVDF membrane by wet-transfer. Membranes were probed with antibodies using standard techniques and detected by enhanced cheiluminscence. RNA polymerase II antibodies: initiating RNA polymerase H14, 1:500 (Covance, MMS-134R) and elongating RNA polymerase H5, 1:500 (Covance, MMS-129R). GAPDH, 1:1000 (Cell Signaling, #2118). (C) Graph showing 30 min pulsed incorporation of 185nM [5-3H] Uridine into short and long RNA species after transcription inhibition. Cold dA, dG, dC, dT and C (37 nM final) were added to cells, to suppress label incorporation into DNA. After 30 min incubation cells were rinsed with PBS and long and short RNAs was extracted by selective binding to a silica matrix (miRNeasy Kit, Qiagen). Residual DNA was removed by on-column DNaseI treatment. RNA was quantified using a Nanodrop and 3H incorporation was measured by scintillation counting.
![Figure 1. Changes in transcription alter DNA supercoiling. (A) Diagram showing inhibition and recovery of transcription after α-amanitin treatment and meta-analysis showing DNA supercoiling around transcription start sites before, during and after transcription inhibition. For methods see reference Citation1. (B) western blot showing levels of RNA polymerase after α-amanitin treatment in the presence and absence of MG132, a proteasome inhibitor. Cells were suspended in 2 x SDS lysis buffer, incubated at 100°C for 5 min and sonicated briefly. Protein samples were resolved on 8% bis-tris gels and transferred to PVDF membrane by wet-transfer. Membranes were probed with antibodies using standard techniques and detected by enhanced cheiluminscence. RNA polymerase II antibodies: initiating RNA polymerase H14, 1:500 (Covance, MMS-134R) and elongating RNA polymerase H5, 1:500 (Covance, MMS-129R). GAPDH, 1:1000 (Cell Signaling, #2118). (C) Graph showing 30 min pulsed incorporation of 185nM [5-3H] Uridine into short and long RNA species after transcription inhibition. Cold dA, dG, dC, dT and C (37 nM final) were added to cells, to suppress label incorporation into DNA. After 30 min incubation cells were rinsed with PBS and long and short RNAs was extracted by selective binding to a silica matrix (miRNeasy Kit, Qiagen). Residual DNA was removed by on-column DNaseI treatment. RNA was quantified using a Nanodrop and 3H incorporation was measured by scintillation counting.](/cms/asset/71737164-4514-4bea-ad74-2ea3a2e41ac0/ktrn_a_10925554_f0001.gif)
As transcription clearly introduced negative DNA supercoils under-winding promoter regions, we investigated further the role of divergent initiation in this process. In our experimental approach, the elongating form of RNA polymerase II (RNAP II) is very sensitive to α-amanitin and is rapidly degraded in a proteasome dependent manner (). In contrast, the initiating form of the polymerase is more resistant to degradation suggesting that changes in supercoiling upon α-amanitin washout are manifest by the initiating form of the polymerase. This was additionally confirmed as blocking transcription elongation specifically with flavopiridol rapidly reduced long RNA production, but there was a lag before a drop in the synthesis of short RNAs, consistent with initiating RNA polymerase producing short RNAs.Citation1 Thus, we reasoned that elongating polymerase synthesizes long RNAs while the initiating form produces shorter RNA species.Citation24 To test this, we trace labeled cells with 3H-Uridine and showed that the production of long RNAs (> 200 nt) was substantially reduced after 5 h α-amanitin treatment and continued to decrease after α-amanitin washout (). In contrast, short RNAs (< 200 nt) were produced in abundance by 1 h recovery and continued to increase at 2 h (), concomitant with remodeling of DNA supercoiling (). We hypothesized that these short RNAs are the products of divergent transcription.Citation16-Citation18 Using data from Core et al.,Citation16 we investigated the expression of several short antisense or sense RNA transcripts around the promoters of the expressed IGBP1 (Xq13.1) and LDHA (11p15.1) genes ().
Figure 2. Short RNA synthesis at TSSs. (A) Diagram showing IGBP1 and LDHA gene loci with sense and anti-sense RNA transcripts. From Core et al.Citation15 (B) Graph showing transcription elongation in the gene-body measured by RT-PCR. Long RNAs (> 200 nt) were reverse transcribed (Superscript II, Invitrogen) using random primers and quantified by qPCR (Fast start SYBR green, Roche). Primer sequences are: IGBP1 Exon1-Intron1: Fwd: ATCTTCAAACCGTGGGAGTG IGBP1 Exon1-Intron1: Rev: AAAACCCTAGGCGCTGTTTT IGBP1 Intron2-Exon3: Few: TTCACTGCCTCCTTTTTGCT IGBP1 Intron2-Exon3: Rev: GCTCAAACTCTGCCACATGA LDHA Intron3-Exon4: Fwd: CAAGAAAGGTTTGTGGAGCA LDHA Intron3-Exon4: Rev: CTTTCTCCCTCTTGCTGACG LDHA Intron2-Exon3: Fwd: AATGGGGTGCCCTCTACTTT LDHA Intron2-Exon3: Rev: AGGCTGCCATGTTGGAGAT (C) Graph showing short RNA transcription measured by miRT-PCR. +/− values show distance from TSS. Short RNAs (< 200 nt) were detected by first poly adenylating and then reverse transcribed using tagged oligo-dT and random primers (miScript kit, Qiagen). They were then quantified using qPCR with a specific forward primer and universal reverse primer (miScript primer assay, Qiagen). Primer sequences are: IGBP1 -277 TTGTCTCTCTACCGCCTTCC IGBP1 -17 GAAGATCCGGTCGCTTGAG LDHA +193 CGATTCCGGATCTCATTG LDHA +279 AGGGATGGGCGGGTAGAG
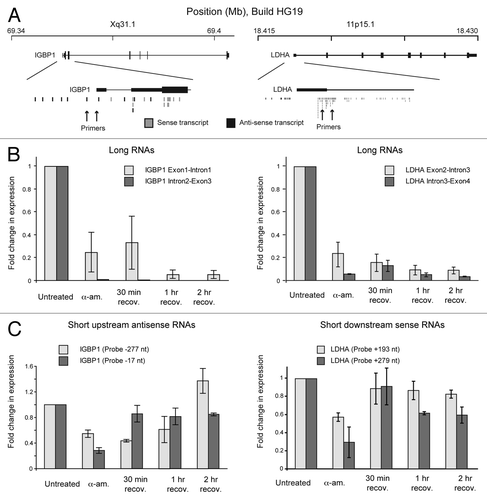
Long RNAs produced within the gene body were inhibited by α-amanitin and did not recover within 2 h (). However, in agreement to what is seen globally (), short antisense and sense RNAs are produced upstream and downstream of genes a short period of time after α-amanitin washout and recovery (). Our data suggests that the initiating form of RNA polymerase, functioning both upstream through divergent transcription and downstream through abortive sense transcription, generates short transcripts and negatively supercoils TSSs creating a permissive environment for subsequent transcription. Furthermore, as DNA supercoiling impacts on higher levels of chromatin organization,Citation1 there is a possible spreading of supercoiling from one locus to another, which could facilitate transcription of surrounding genes and might provide a rationale for gene clustering in the human genome.
This model harmonizes recent advances in our understanding of transcriptional regulation. Traditionally, RNAP II recruitment is thought to be the rate limiting step and, thus, the key regulatory step in eukaryotic transcription; however, genome wide profiling of RNAP II indicates that it is bound and initiated at both active and inactive genes.Citation25 Indeed, for a large proportion of metazoan genes (20–30%), RNAP II density is enriched downstream of many TSSs and this has been described as RNAP II promoter proximal pausing.Citation26,Citation27 Pausing is now thought to be a widespread regulatory mechanism with the Negative Elongation Factor (NELF) and DRB-Sensitivity Inducing Factor (DSIF) protein complexes binding to and arresting RNAP II 60 nts downstream of the TSS. Subsequent recruitment of Positive Transcription Elongation Factor b (P-TEFb) to this paused RNAP II complex and phosposphorylation of DSIF, NELF and Ser2 on the RNAP II C-terminal results in dissociation of NELF and productive transcriptional elongation. Both sense and antisense RNAP II complexes are involved in RNAP II pausing and both depend on PTEF-b recruitment.Citation19
The purpose of pausing is not known but it is frequently found at developmental control genes and stimulus-responsive pathways and is though to allow their rapid and synchronous induction in response to extracellular signals.Citation27 Consequently, loss of pausing through knockdown of the pause-inducing factor NELF leads to broadly attenuated immune gene activation.Citation28 One function of paused RNAP II is to establish a permissive chromatin environmentCitation29 and paused polymerase has been shown to block nucleosome assembly at promoters, thus maintaining an open chromatin architecture.Citation20,Citation30 In support of this, we propose that promoter proximal pausing permits time for divergent transcription to produce short RNAs and, concomitantly, negatively supercoil promoter regions to facilitate transcription. After transcriptional pause-release, polymerases in collaboration with topoisomerase and helicases can then maintain the supercoiling state of the locus in a regulated manner. This is consistent with pausing being more frequently associated with developmentally regulated genes, while constitutively expressed genes are maintained with optimal levels of DNA supercoiling. However, anti-sense RNA transcription is significantly less prevalent in Drosophila,Citation24,Citation31 but there is pronounced promoter pausing. This may suggest that the purpose of pausing may be different in mammalian and Drosophila genomes, or that abortive sense transcription is sufficient to negatively supercoil the promoter to facilitate transcription factor binding and subsequent processive transcription. The fact that no function has as yet been assigned to the short sense and antisense RNAs further substantiates our idea that they are by products of a critical process necessary to create a transcriptionally friendly chromatin environment. Future work combining targeted RNAi of RNAP II pausing and elongation factors with DNA supercoiling analysis will elucidate the mechanism by which chromatin structure influences transcription.
Acknowledgments
This work was funded by the Wellcome Trust 078219/Z/05/Z and N.G. is a recipient of a UK Medical Research Council senior fellowship (MR/J00913X/1).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Naughton C, Avlonitis N, Corless S, Prendergast JG, Mati IK, Eijk PP, et al. Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat Struct Mol Biol 2013; 20:387 - 95; http://dx.doi.org/10.1038/nsmb.2509; PMID: 23416946
- Kouzine F, Gupta A, Baranello L, Wojtowicz D, Ben-Aissa K, Liu J, et al. Transcription-dependent dynamic supercoiling is a short-range genomic force. Nat Struct Mol Biol 2013; 20:396 - 403; http://dx.doi.org/10.1038/nsmb.2517; PMID: 23416947
- Matsumoto K, Hirose S. Visualization of unconstrained negative supercoils of DNA on polytene chromosomes of Drosophila. J Cell Sci 2004; 117:3797 - 805; http://dx.doi.org/10.1242/jcs.01225; PMID: 15252118
- Kouzine F, Sanford S, Elisha-Feil Z, Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat Struct Mol Biol 2008; 15:146 - 54; http://dx.doi.org/10.1038/nsmb.1372; PMID: 18193062
- Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A 1987; 84:7024 - 7; http://dx.doi.org/10.1073/pnas.84.20.7024; PMID: 2823250
- Stewart AF, Herrera RE, Nordheim A. Rapid induction of c-fos transcription reveals quantitative linkage of RNA polymerase II and DNA topoisomerase I enzyme activities. Cell 1990; 60:141 - 9; http://dx.doi.org/10.1016/0092-8674(90)90724-S; PMID: 2153054
- Gilmour DS, Pflugfelder G, Wang JC, Lis JT. Topoisomerase I interacts with transcribed regions in Drosophila cells. Cell 1986; 44:401 - 7; http://dx.doi.org/10.1016/0092-8674(86)90461-7; PMID: 3002635
- Harland RM, Weintraub H, McKnight SL. Transcription of DNA injected into Xenopus oocytes is influenced by template topology. Nature 1983; 302:38 - 43; http://dx.doi.org/10.1038/302038a0; PMID: 6828158
- Weintraub H, Cheng PF, Conrad K. Expression of transfected DNA depends on DNA topology. Cell 1986; 46:115 - 22; http://dx.doi.org/10.1016/0092-8674(86)90865-2; PMID: 3459589
- Mizutani M, Ohta T, Watanabe H, Handa H, Hirose S. Negative supercoiling of DNA facilitates an interaction between transcription factor IID and the fibroin gene promoter. Proc Natl Acad Sci U S A 1991; 88:718 - 22; http://dx.doi.org/10.1073/pnas.88.3.718; PMID: 1992462
- Parvin JD, Sharp PA. DNA topology and a minimal set of basal factors for transcription by RNA polymerase II. Cell 1993; 73:533 - 40; http://dx.doi.org/10.1016/0092-8674(93)90140-L; PMID: 8490964
- Goodrich JA, Tjian R. Transcription factors IIE and IIH and ATP hydrolysis direct promoter clearance by RNA polymerase II. Cell 1994; 77:145 - 56; http://dx.doi.org/10.1016/0092-8674(94)90242-9; PMID: 8156590
- Ljungman M, Hanawalt PC. Localized torsional tension in the DNA of human cells. Proc Natl Acad Sci U S A 1992; 89:6055 - 9; http://dx.doi.org/10.1073/pnas.89.13.6055; PMID: 1631091
- Ljungman M, Hanawalt PC. Presence of negative torsional tension in the promoter region of the transcriptionally poised dihydrofolate reductase gene in vivo. Nucleic Acids Res 1995; 23:1782 - 9; http://dx.doi.org/10.1093/nar/23.10.1782; PMID: 7784183
- Dunaway M, Ostrander EA. Local domains of supercoiling activate a eukaryotic promoter in vivo. Nature 1993; 361:746 - 8; http://dx.doi.org/10.1038/361746a0; PMID: 8441472
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 2008; 322:1845 - 8; http://dx.doi.org/10.1126/science.1162228; PMID: 19056941
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, et al. Divergent transcription from active promoters. Science 2008; 322:1849 - 51; http://dx.doi.org/10.1126/science.1162253; PMID: 19056940
- Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, et al. RNA exosome depletion reveals transcription upstream of active human promoters. Science 2008; 322:1851 - 4; http://dx.doi.org/10.1126/science.1164096; PMID: 19056938
- Flynn RA, Almada AE, Zamudio JR, Sharp PA. Antisense RNA polymerase II divergent transcripts are P-TEFb dependent and substrates for the RNA exosome. Proc Natl Acad Sci U S A 2011; 108:10460 - 5; http://dx.doi.org/10.1073/pnas.1106630108; PMID: 21670248
- Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev 2011; 25:742 - 54; http://dx.doi.org/10.1101/gad.2005511; PMID: 21460038
- Wei W, Pelechano V, Järvelin AI, Steinmetz LM. Functional consequences of bidirectional promoters. Trends Genet 2011; 27:267 - 76; http://dx.doi.org/10.1016/j.tig.2011.04.002; PMID: 21601935
- Seila AC, Core LJ, Lis JT, Sharp PA. Divergent transcription: a new feature of active promoters. Cell Cycle 2009; 8:2557 - 64; http://dx.doi.org/10.4161/cc.8.16.9305; PMID: 19597342
- Sinden RR, Bat O, Kramer PR. Psoralen cross-linking as probe of torsional tension and topological domain size in vivo. Methods 1999; 17:112 - 24; http://dx.doi.org/10.1006/meth.1998.0723; PMID: 10075890
- Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science 2010; 327:335 - 8; http://dx.doi.org/10.1126/science.1181421; PMID: 20007866
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 2007; 130:77 - 88; http://dx.doi.org/10.1016/j.cell.2007.05.042; PMID: 17632057
- Gilmour DS, Lis JT. RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol Cell Biol 1986; 6:3984 - 9; PMID: 3099167
- Li J, Gilmour DS. Promoter proximal pausing and the control of gene expression. Curr Opin Genet Dev 2011; 21:231 - 5; http://dx.doi.org/10.1016/j.gde.2011.01.010; PMID: 21324670
- Gilchrist DA, Fromm G, dos Santos G, Pham LN, McDaniel IE, Burkholder A, et al. Regulating the regulators: the pervasive effects of Pol II pausing on stimulus-responsive gene networks. Genes Dev 2012; 26:933 - 44; http://dx.doi.org/10.1101/gad.187781.112; PMID: 22549956
- Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet 2012; 13:720 - 31; http://dx.doi.org/10.1038/nrg3293; PMID: 22986266
- Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, et al. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell 2010; 143:540 - 51; http://dx.doi.org/10.1016/j.cell.2010.10.004; PMID: 21074046
- Core LJ, Waterfall JJ, Gilchrist DA, Fargo DC, Kwak H, Adelman K, et al. Defining the status of RNA polymerase at promoters. Cell Rep 2012; 2:1025 - 35; http://dx.doi.org/10.1016/j.celrep.2012.08.034; PMID: 23062713