Abstract
Post-transcriptional control of mRNA is a vital step in the regulation of gene expression. Highly specific combinations of RNA-binding–proteins (RBPs) and microRNAs conjointly contribute to the complexity of post-transcriptional control of mRNAs. Post-transcriptional switches regulated by RBPs control context-specific expression of two alternative gene products from a single transcript.
In the pathway from transcription to translation, RNA was generally considered as a passive molecule. The structural components of mRNAs usually consist of a 5′- m7GpppN cap, a 5′-untranslated region (UTR), the coding region, a 3′-UTR and a poly(A) tail. Initiation of translation occurs by a sequence of events starting with the binding of a cap-binding complex, recruitment of the pre-initiation complex with 40S ribosomal subunit, scanning and, finally, recruitment of the 60S large ribosomal subunit.Citation1 Adding to the intricacy, interaction of RNA-binding proteins (RBPs) with 5′- or 3′-UTRs of mRNA and sequence-specific binding of microRNAs (miRNAs), together control the efficiency of translation of the transcript.
Transcripts and cis-Regulatory modules
Transcript-specific translational control of eukaryotic gene expression is often directed by a structural element in the 3′-UTR of the mRNA.Citation2 In most vertebrates, 3′-UTRs are substantially longer than their 5′ counterparts, suggesting a significant potential for regulation. Furthermore, a multi-species analysis indicates an increase in the average length of 3′-UTR sequences during evolution, highlighting a correlation of these sequences with increasing complexity of the organism.Citation3 Several protein-coding genes contain miRNA binding sites within their 3′-UTRs. miRNAs are small, non-coding RNAs that influence diverse biological functions through the repression of target genes. It has been proposed that complete complementarity between miRNA and target mRNA leads to the degradation of target mRNA,Citation4 whereas partial complementarity may lead to translational repression and/or mRNA degradation.Citation5
MicroRNAs can be classified broadly into three categories: intergenic, intronic and exonic, based on genomic location, coordinates, and source of primary transcript.Citation6 Although 52% of human miRNAs are intergenic, ~43% of the remaining human miRNAs are encoded within the host genes. One notable feature of intragenic miRNAs is the co-regulation of both miRNAs as well as host mRNA with a single transcription factor/pathway. The remaining ~5% of miRNAs are encoded from the exons of protein coding genes. They are referred to as exonic miRNAs, characterized by the presence of precursor-miRNA stem loop within the exon of the transcripts.Citation7 The first exonic miRNA identified was miR-675 derived from the first exon of H19 non-coding RNA.Citation8 miR-675 is highly expressed in placenta. In majority of the tissues miR-675 processing from H19 long non-coding RNA is suppressed by an RBP, HuR.
A Repertoire of RBPs Coordinate Multiple Post-transcriptional Events
RBPs interact with regulatory elements of transcripts and control spatial and tissue-specific expression of proteins.Citation3 After transcription, the fate of the transcript is determined by a complex interplay of RBPs and regulatory elements within the transcripts. In multiple instances, RNA stabilizing RBPs like HuR inhibit miRNA mediated repression/deadenylation of target mRNAs by dissociating miRISC/Ago complex from target mRNAs.Citation9 HuR promotes translation of nucleolin mRNA in HeLa cells. However, in the absence of HuR, miR-494 translocates nucleolin mRNA to processing bodies (P bodies).Citation10 Some of the multifunctional RBPs also play a vital role in the biogenesis/processing of miRNAs. Trabucchi et al. detected the association of KH-type splicing regulator protein (KSRP) with the Dicer complex, suggesting a potential role for KSRP in miRNA biogenesis.Citation11 Interestingly, processing of a single miRNA may be controlled by multiple developmentally regulated proteins. Let-7 is regulated by Lin28 in embryonic cells, whereas KSRP promotes the biogenesis of this miRNA in differentiated cells. Paradoxically, another RBP, hnRNPA1, competes with KSRP in binding to the terminal loop of pri-let-7 and inhibits let-7 processing.Citation12,Citation13 KSRP-mediated miR-155 biogenesis is also inhibited by an RBP Tristetraproline (TTP) in fibrotic lung epithelial cells, although the exact mechanism of this inhibition is unclear.Citation14 Furthermore, KSRP interacts with both the primary-miRNA transcript and the stem-loop precursor through a G-rich motif present in a subset of mRNAs and promote miRNA biogenesis.Citation11
An Inter-play Between RBPs and Growth Factors Control Post-transcriptional Switches
An intricate network of secreted factors and intracellular signaling intermediates control robust gene expression by cross-talks or direct interaction with RBPs. Growth factors, such as transforming growth factor-β (TGF-β) involved in signaling pathways, modulate the activities of the components of the miRNA biogenesis and indirectly control miRNA expression patterns.Citation15 The major elements in TGF-β signaling are TGF-β ligands, their receptors, and intracellular Smad effectors. Signal transducers of the TGF-β pathway, the Smad proteins, also modulate miRNA expression by transcriptional and post-transcriptional mechanisms.Citation16 TGF-β and Bone Morphogenetic Protein 4 (BMP4) signaling stimulate the production of pre-miR-21 by facilitating Drosha-mediated processing. Nuclear accumulation of receptor-Smads (R-Smads) proteins, regulate miRNA maturation by associating with the Drosha/DGCR8 complex.Citation17
Highlighting some of the above-mentioned events we describe an elegant post-transcriptional switch that controls the context-specific expression of miR-198 or follistatin-like-1 (FSTL1) from a single transcript.Citation18 miR-198 is a primate-specific miRNA and belongs to a small cohort of human exonic miRNA stem–loops located within the eleventh exon of the protein-coding gene, FSTL1.Cutaneous epidermal keratinocytes express high levels of miR-198. Targeting multiple genes including urokinase-type plasminogen activator (PLAU), diaphanous homolog 1 (DIAPH1), and laminin gamma 2 chain (LAMC2), essential for migration, miR-198 inhibits keratinocyte migration. However, upon skin injury, miR-198 levels plummet as early as 3 hours after wounding, resulting in expression of the target genes and migration of keratinocytes toward the wound edge. Most importantly, expression of miR-198 from the 3′-UTR of FSTL1 mRNA in normal skin switches to expression of the linked open reading frame (ORF) of FSTL1 upon wounding. Expression of glycoprotein FSTL1 was clearly observed at the wound edge, but not in unwounded normal epidermal keratinocytes. Unlike miR-198, FSTL1 facilitates keratinocyte migration. A clear inverse correlation exists between the expression pattern of FSTL1, essential for effective migration and miR-198, an inhibitor of migration.Citation18 A single transcript can therefore function either as a primary miRNA transcript making miR-198 or function as an mRNA producing FSTL1 protein, suggesting a post-transcriptional switch regulating their context-specific expressionCitation18,Citation19 ().
Figure 1. Schematic representation of a post-transcriptional switch controlling context-specific expression of two alternative gene products.
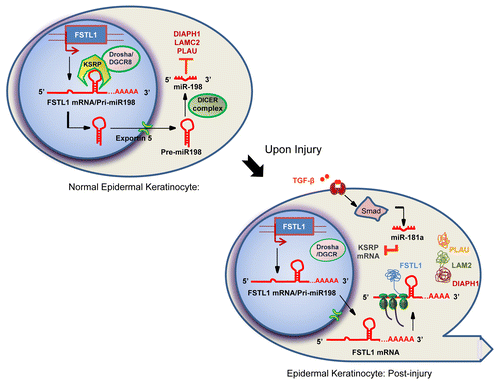
The mere presence of a miRNA precursor stem–loop in cis may not result in the processing of the miRNA. Adding another layer of complexity, miR-198 belongs to a small cohort of miRNAs that require KSRP for processing. KSRP binds to the GUG motif in the terminal loop of - miR-198 stem-loop precursor within the 3′-UTR of FSTL1 transcript and facilitates processing of miR-198. In the absence of KSRP, the transcript functions as an mRNA making the protein FSTL1, highlighting the essential role of KSRP in switching-on miR-198 processing.Citation18
However, upon injury, TGF-β signaling turns-off miR-198 processing by down-regulating KSRP and promotes FSTL1 expression. TGF-β down-regulates KSRP through another miRNA, miR-181a. In summary, this complex network regulated by TGF-β and KSRP controls context-specific expression of two alternative gene products from a single transcript that functions either as an mRNA or as a pri-miRNA transcriptCitation18 (Fig.. 1).
Post-transcriptional Control and its Implications in Physiology and Pathophysiology of Diseases
It has been demonstrated that perturbations in RBPs may consequently lead to profound phenotypic alterations in homeostasis resulting in a pathological condition. RBPs and miRNAs co-operatively regulate multiple disease pathways, indicating complementarity between the two types of post-transcriptional regulation. Hereditary hyperferritinemia-cataract syndrome (HHCS) is characterized by bilateral cataracts and increased serum L-ferritin, in the absence of iron overload. Regulation of iron metabolism occurs mainly at the post-transcriptional level. Trans-acting iron regulatory proteins bind to the cis-acting iron-responsive elements, located in the UTRs of target mRNAs and control transcript levels.Citation20 Similarly, regulation of neurofilament protein expression depends on posttranscriptional control of its mRNA including localization, translational efficiency and stability. Imbalances in the expression may lead to formation of neurofilament aggregates resulting in neurodegenerative diseases.Citation21 Cyclo-oxygenase-2 (COX-2), aberrantly expressed in multiple stages of colon carcinogenesis, is involved in the synthesis of prostaglandins. In normal intestinal epithelium, this message is degraded by the action of miR-16. However, in colorectal cancers, the mRNA stability factor HuR inhibits miR-16 targeting of COX-2.Citation22 Similarly, in prostate cancer, HuR opposes the repression of ERBB-2 gene expression by miR-331–3p.Citation23
Adding to this list, we demonstrate a non-functional miR-198/FSTL1 post-transcriptional switch in chronic diabetic ulcer wounds from patients with diabetes mellitus, where wound healing is defective. We found persistent high levels of miR-198 at the chronic wound edge. Furthermore, absence of FSTL1 protein at the wound edge indicates a defective switch in chronic wounds.Citation18 In conclusion, a non-functional switch leads to impaired keratinocyte migration and loss of re-epithelialization culminating in non-healing chronic wounds, a consequence of defective TGF-β signaling. In patients with diabetes mellitus, chronic wounds are a major health burden resulting in lower extremity amputationsCitation24 and modulation of the defective switch may lead to wound re-epithelialization and effective wound healing, resulting in improved patient outcome.
Conclusions
Extensive work on post-transcriptional control of eukaryotic gene expression indicates that mRNA translation is influenced by an array of regulatory mechanisms. The impact of post-transcriptional switches in the dynamic regulation of proteome is increasingly appreciated. Cell-type specific post-transcriptional switches modulate gene expression by multiple modes. These regulatory switches are under the stringent control of growth factors, RBPs and miRNAs. It is challenging to predict the mechanisms involved in the processing of exonic miRNAs. If the mRNA harbors the precursor miRNA stem-loop in its UTR, whether the transcript encodes a functional protein or gets processed into a miRNA is still an open question for multiple exonic miRNAs. Additionally, it would be interesting to see if such a transcript can coordinately express both the encoded protein and the linked miRNA. Complicating the scenario, such post-transcriptional switches are regulated by an intricate network of RBPs and growth factors with extensive cross-talk. Unraveling these networks and elucidating novel post-transcriptional switches may provide an answer for the aberrant gene expression programs in multiple pathological conditions.
Acknowledgments
This work was supported by an A*STAR Investigatorship award to PS, Biomedical Research Council of Singapore and the Skin Biology Cluster Platform, A*STAR.
Submitted
07/14/13
Revised
09/03/13
Accepted
09/05/13
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol 2004; 5:827 - 35; http://dx.doi.org/10.1038/nrm1488; PMID: 15459663
- Sampath P, Mazumder B, Seshadri V, Fox PL. Transcript-selective translational silencing by gamma interferon is directed by a novel structural element in the ceruloplasmin mRNA 3′ untranslated region. Mol Cell Biol 2003; 23:1509 - 19; http://dx.doi.org/10.1128/MCB.23.5.1509-1519.2003; PMID: 12588972
- Mazumder B, Seshadri V, Fox PL. Translational control by the 3′-UTR: the ends specify the means. Trends Biochem Sci 2003; 28:91 - 8; http://dx.doi.org/10.1016/S0968-0004(03)00002-1; PMID: 12575997
- Tang G, Reinhart BJ, Bartel DP, Zamore PD. A biochemical framework for RNA silencing in plants. Genes Dev 2003; 17:49 - 63; http://dx.doi.org/10.1101/gad.1048103; PMID: 12514099
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 2004; 5:522 - 31; http://dx.doi.org/10.1038/nrg1379; PMID: 15211354
- Hinske LC, Galante PA, Kuo WP, Ohno-Machado L. A potential role for intragenic miRNAs on their hosts’ interactome. BMC Genomics 2010; 11:533; http://dx.doi.org/10.1186/1471-2164-11-533; PMID: 20920310
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 2009; 10:126 - 39; http://dx.doi.org/10.1038/nrm2632; PMID: 19165215
- Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, Reik W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol 2012; 14:659 - 65; http://dx.doi.org/10.1038/ncb2521; PMID: 22684254
- Kundu P, Fabian MR, Sonenberg N, Bhattacharyya SN, Filipowicz W. HuR protein attenuates miRNA-mediated repression by promoting miRISC dissociation from the target RNA. Nucleic Acids Res 2012; 40:5088 - 100; http://dx.doi.org/10.1093/nar/gks148; PMID: 22362743
- Tominaga K, Srikantan S, Lee EK, Subaran SS, Martindale JL, Abdelmohsen K, Gorospe M. Competitive regulation of nucleolin expression by HuR and miR-494. Mol Cell Biol 2011; 31:4219 - 31; http://dx.doi.org/10.1128/MCB.05955-11; PMID: 21859890
- Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature 2009; 459:1010 - 4; http://dx.doi.org/10.1038/nature08025; PMID: 19458619
- Guil S, Cáceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol 2007; 14:591 - 6; http://dx.doi.org/10.1038/nsmb1250; PMID: 17558416
- Michlewski G, Cáceres JF. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat Struct Mol Biol 2010; 17:1011 - 8; http://dx.doi.org/10.1038/nsmb.1874; PMID: 20639884
- Bhattacharyya S, Kumar P, Tsuchiya M, Bhattacharyya A, Biswas R. Regulation of miR-155 biogenesis in cystic fibrosis lung epithelial cells: antagonistic role of two mRNA-destabilizing proteins, KSRP and TTP. Biochem Biophys Res Commun 2013; 433:484 - 8; http://dx.doi.org/10.1016/j.bbrc.2013.03.025; PMID: 23524258
- Ikushima H, Miyazono K. TGF-β signal transduction spreading to a wider field: a broad variety of mechanisms for context-dependent effects of TGF-β. Cell Tissue Res 2012; 347:37 - 49; http://dx.doi.org/10.1007/s00441-011-1179-5; PMID: 21618142
- Davis BN, Hata A. Regulation of MicroRNA Biogenesis: A miRiad of mechanisms. Cell Commun Signal 2009; 7:18; http://dx.doi.org/10.1186/1478-811X-7-18; PMID: 19664273
- Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature 2008; 454:56 - 61; http://dx.doi.org/10.1038/nature07086; PMID: 18548003
- Sundaram GM, Common JE, Gopal FE, Srikanta S, Lakshman K, Lunny DP, Lim TC, Tanavde V, Lane EB, Sampath P. ‘See-saw’ expression of microRNA-198 and FSTL1 from a single transcript in wound healing. Nature 2013; 495:103 - 6; http://dx.doi.org/10.1038/nature11890; PMID: 23395958
- Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 2004; 10:1957 - 66; http://dx.doi.org/10.1261/rna.7135204; PMID: 15525708
- Mikulits W, Schranzhofer M, Beug H, Müllner EW. Post-transcriptional control via iron-responsive elements: the impact of aberrations in hereditary disease. Mutat Res 1999; 437:219 - 30; http://dx.doi.org/10.1016/S1383-5742(99)00085-X; PMID: 10592329
- Thyagarajan A, Strong MJ, Szaro BG. Post-transcriptional control of neurofilaments in development and disease. Exp Cell Res 2007; 313:2088 - 97; http://dx.doi.org/10.1016/j.yexcr.2007.02.014; PMID: 17428473
- Young LE, Moore AE, Sokol L, Meisner-Kober N, Dixon DA. The mRNA stability factor HuR inhibits microRNA-16 targeting of COX-2. Mol Cancer Res 2012; 10:167 - 80; http://dx.doi.org/10.1158/1541-7786.MCR-11-0337; PMID: 22049153
- Epis MR, Barker A, Giles KM, Beveridge DJ, Leedman PJ. The RNA-binding protein HuR opposes the repression of ERBB-2 gene expression by microRNA miR-331-3p in prostate cancer cells. J Biol Chem 2011; 286:41442 - 54; http://dx.doi.org/10.1074/jbc.M111.301481; PMID: 21971048
- Brem H, Tomic-Canic M. Cellular and molecular basis of wound healing in diabetes. J Clin Invest 2007; 117:1219 - 22; http://dx.doi.org/10.1172/JCI32169; PMID: 17476353