Abstract
Most RNA polymerases contain zinc, yet the precise function of zinc and its influence of polymerases stability are unknown. A recent study provides evidence that zinc levels control the stability of RNA polymerase I in vivo and that the enzyme might serve as a zinc reservoir for other proteins.
Background: Zinc Binding and Zinc-dependent Activity of Eukaryotic RNA Polymerases
The three major eukaryotic RNA polymerases contain zinc. The presence of zinc cations bound to various polymerase subunits was determined by early work describing the purification of eukaryotic RNA polymerasesCitation1-Citation5, including RNA polymerase I (RNAP I), the enzyme that produces three of the four rRNAs. These pioneer studies also showed that zinc is required for the activity of various eukaryotic RNA polymerases, as treatment with zinc-specific chelators result in a specific inhibition of the polymerase activity. Other approaches, including zinc blotting experiments, subsequently identified subunits within the RNAP I complex and within other eukaryotic polymerases that bind zinc very tightlyCitation5. While it was clear from these early studies that zinc is required for eukaryotic RNA polymerase activity in vitro, the influence of zinc on the activity of RNA polymerases in conditions in which organisms grow in the absence of zinc or with a very limited zinc supply (zinc deficiency) has remained poorly understood. Early work from B. Vallée's group showed that Euglena grown in conditions of zinc deficiency showed only a single peak of RNA polymerase activity, which is likely to correspond to an α amanitin-resistant form of RNA polymerase II (RNAP II)Citation6. The precise identity of this polymerase, as well as the cellular mechanisms that result in this single peak were never identified to our knowledge. In mammalian cells, work on rat embryos showed that zinc deficiency resulted in a decrease in overall RNA polymerase activityCitation7. However, it was not clear from this work which of the RNA polymerase contributed the most to the bulk activity monitored in that assay, and whether each of the three major RNA polymerases was affected individually to the same extent. In the yeast S.cerevisiae, zinc deficiency initiates a transcriptional response mediated by RNAP II that is responsible for the expression of specific genes required to respond to zinc starvationCitation8. Multiple studies have shown that the transcriptional activator Zap1p mediates the activation of genes needed for survival in low zinc conditionsCitation8-Citation10. However, the transcriptional activities of the other two RNA polymerases were never fully characterized in these conditions. In a recent study, we showed that in addition to the requirement of zinc for RNAP I activity in vitro, zinc deficiency directly affects the stability of the RNAP I complex in vivo by triggering its export from the nucleus and degradation in the vacuole. The mechanisms and biological significance of this downregulation are further discussed below.
Vacuolar Autophagy of RNA Polymerase I During Zinc Deficiency
In a recent studyCitation11, we showed that in conditions of zinc deficiency, S.cerevisiae RNAP I and a some of its associated proteins are downregulated by nuclear export to the vacuole and degradation by vacuolar proteases (). Vacuolar export and degradation of RNAP I is specific to zinc deficiency, as it is not observed when cells are deprived of other transition metals like iron, or in other stress conditions such as amino acid starvation. In addition, RNAP II is not affected, consistent with numerous studies showing activated transcription by RNAP II of genes involved in the response to zinc deficiencyCitation8-Citation10. Surprisingly, some proteins found associated with RNAP I are also degraded in the vacuole, including some rRNA processing factors. This observation suggests that export of the RNAP I subunits occurs within the context of the core RNAP I complex and its associated proteinsCitation12. Export out of the nucleus requires the general export factor Xpo1p () but the detailed mechanisms of the subsequent import of the RNAP I into the vacuole remain poorly understood. Proteins are typically imported in the yeast vacuole (the functional equivalent of mammalian cells lysosome) through autophagy or cytoplasm to vacuole (CVT) targeting pathwaysCitation13. However, mutants deficient in these targeting pathways did not show an inhibition of RNAP I downregulation, suggesting that the import of RNAP I in the vacuole does not require these classical pathways. In addition, vacuolar import of RNAP I does not involve the piecemeal microautophagy of the nucleus (PMN) pathway which has been shown to induce the formation of nuclear-vacuole junctions in stress conditionsCitation14. Thus the specific molecular mechanisms by which RNAP I is imported to the vacuole after it is export out of the nucleus remain to be identified.
Figure 1. Shown is the equilibrium between ubiquitinated states of the RNA polymerase I complex, and the consequences of these differences in ubiquitination on the stability of the Polymerase I complex. The identity of the ubiquitin proteases that promote the deubiquitination reactions is indicated. Ubi1 and Ubi2 would correspond to the different ubiquitinated states, which would result in RNAP I degradation and nuclear retention, respectively.
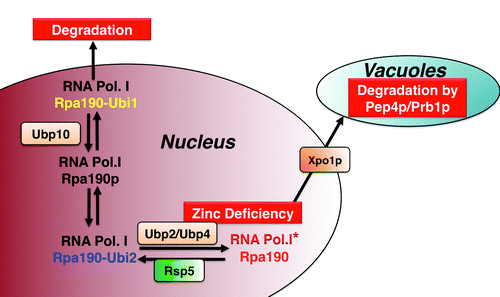
Dual Role for Ubiquitination in the Stability of RNA Polymerase
The mechanism that triggers vacuolar export and degradation of RNAP I provides an interesting perspective on the antagonistic functions of ubiquitination on RNAP I stability. Recent work from Richardson et al.Citation15 showed that ubiquitination of the large subunit Rpa190p destabilizes RNAP I in normal growth conditions (), and that inactivation of the Ubp10p ubiquitin protease is sufficient to decrease RNAP I levels, resulting in a decrease in rRNA biogenesis. By contrast, during zinc deficiency, a global deubiquitination of the RNAP I large subunit Rpa190p can be detected. The activity of the Ubp2p and Ubp4p ubiquitin proteases was found to be required for vacuolar export of RNA polymerase I, as inactivation of Ubp2p and Ubp4p results in a stabilization of RNAP I in zinc deficiency and in the inhibition of vacuolar import. This result is consistent with a role for deubiquitination in signaling export out of the nucleus and/or vacuolar import (), hinting to a possible role for Ubp2p/Ubp4p in the deubiquitination of Rpa190p that triggers nuclear export. However, it remains to be established whether Ubp2p and/or Ubp4p directly deubiquitinate Rpa190p in zinc deficiency. The suppression of RNAP I degradation in the Ubp2p and Ubp4p mutants could be indirect, as these proteins have a general role in the import of proteins into the vacuoleCitation16.
The apparent contradictory effects of ubiquitination on the stability of RNAP I in normal conditions compared with zinc deficiency could be explained by the presence of multiple, yet functionally distinct ubiquitination sites on the large subunit Rpa190p (). Ubiquitination at some of these sites would be regulated by Ubp10pCitation15 and would lead to degradation, presumably by the proteasome system, while ubiquitination at other sites would prevent nuclear export of RNAP I. In addition, it seems likely that the deubiquitination of RNAP I that is promoted by zinc deficiency should result in a complex that is structurally different from the deubiquitinated RNP I that results in its stabilization in normal zinc conditions (). Thus, a full understanding of how ubiquitination modulates the stability of the RNAP I complex will require characterization of different ubiquitination sites and their respective functions in promoting RNAP I degradation or nuclear retention.
Has Zinc Binding Evolved in RNA Polymerase I to Serve as a Zinc Reservoir?
Why did S.cerevisiae cells evolve a mechanism to degrade the RNAP I complex during zinc deficiency? The precise function of the downregulation of RNAP I during zinc deficiency is difficult to address experimentally. Transcription of the rDNA by RNAP I uses a massive amount of nucleotides, and the rRNA generated by this transcription is then packaged into ribosomes. Ribosome biogenesis itself is also extremely costly energeticallyCitation17. Thus, at first glance, the downregulation of RNAP I during zinc deficiency might seem to be an austerity response to the limited resources available during zinc deficiency. By downregulating ribosome biogenesis, yeast cells would focus their limited metabolic resources to other functions that are more important for survival in low zinc conditions. However there are easier and less drastic ways to regulate ribosome biogenesis than to degrade the entire RNAP I complex. For instance, many stress conditions are known to downregulate rDNA transcription, usually by modulating the number of RNAP I subunits associated with the transcription factors that drive its association to the rDNA promoterCitation18. This type of regulatory mechanism does not affect the steady-state level of the polymerase, and one of the advantages of this type of regulation is that cells can reinitiate rRNA synthesis quickly when cells are exposed back to normal growth conditions. Thus, it is likely that the downregulation of RNAP I has evolved to serve a different purpose than simply downregulating ribosome biogenesis.
We proposed that the degradation of RNAP I in the vacuole might serve as a means for cells to recover some of the zinc bound to RNAP I subunits, and to redistribute these zinc atoms to other proteins whose functions are strictly required in zinc deficiency. This hypothesis is based on the fact that redistribution of metals from protein to other proteins has been described in conditions of metal deficiency, and also because the vacuole, the organelle where RNAP I is degraded is also the site of storage and redistribution of most metals, including zinc in yeastCitation19. Thus, vacuolar degradation of the zinc-binding subunits would allow the recovery of zinc bound to these subunits. Given an estimated number of 5,000 RNAP I molecules per cell, and an estimated number of 10 zinc atoms per complex, degradation of the Pol. I complex might be sufficient to increase globally the cellular zinc concentration by approximately 2.7µM. The hypothesis that RNAP I might constitute a cellular zinc reservoir is supported by the observation that ubiquitin protease mutant strains that impair vacuolar import of RNAP I exhibit a growth defect in zinc-deficient medium. Thus, import of RNAP I to the vacuole seems to provide a selective advantage to cells in conditions of zinc-deficiency. Despite this observation, we cannot rule out that the growth defect of these mutants in zinc-deficient medium is due to indirect effects, rather than to the inability to import and degrade RNAP I in the vacuole. The unambiguous demonstration that RNAP I degradation provides a last resort zinc supply to cells starved for zinc would require the characterization of RNAP I subunit mutants that can no longer be imported in the vacuole, yet retain functionality in rDNA transcription. If these mutants result in a specific growth defect in zinc deficiency, they would demonstrate unambiguously that the ability to import RNAP I in the vacuole is important for cellular fitness, likely because of the requirement to recycle the zinc atoms.
Perspective and Future Directions
The discovery that Polymerase I is exported and degraded in the vacuole in conditions of zinc starvation leads to several future directions. Whether this mechanism is unique to yeast or conserved in other eukaryotic cells remains to be determined. Earlier work on rats fed in a zinc deficient diet showing lowered global RNA polymerase activityCitation7, and the observation that a single enzyme similar to RNAP II can be detected in Euglena in zinc deficiencyCitation6 suggests that this physiological response might be conserved across eukaryotes. In addition, the identity of the zinc binding sites in the various RNAP I subunits and the potential zinc lability on these sites in conditions of zinc deficiency remain to be established. One can speculate that some of these sites might release zinc very quickly, thus serving as a molecular signal to trigger nuclear export. The characterization of zinc-binding mutants in various RNAP I subunits might reveal the precise functions of specific zinc binding sites in zinc economy. This, however, is complicated by the observation that some of the sites that are presumed to bind zinc do not seem to be involved in zinc binding, but rather are important to promote assembly of the polymeraseCitation20. Finally it remains to be determined what molecular signal triggers the deubiquitination and export of RNAP I out of the nucleolus during zinc deficiency. Whether a decrease in cellular zinc levels directly influences the activity of the ubiquitin-conjugating enzymes/deubiquitinases or triggers a cascade that ultimately controls these enzymes remains to be determined. Regardless of the precise mechanisms, the observation that RNAP I stability can be tightly controlled by zinc levels opens the door to possible therapeutic manipulation of Polymerase I stability, as this enzyme has emerged as a potential target for cancer treatmentCitation21.
Acknowledgments
I thank K. Roy for critical reading of the manuscript. Work in the author's group is supported by grant R01 GM61518 from the NIGMS.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Auld DS, Atsuya I. Yeast RNA polymerase I: a eukaryotic zinc metalloenzyme. Biochem Biophys Res Commun 1976; 69:548 - 54; http://dx.doi.org/10.1016/0006-291X(76)90555-6; PMID: 773379
- Falchuk KH, Mazus B, Ulpino L, Vallee BL. Euglena gracilis DNA dependent RNA polymerase II: a zinc metalloenzyme. Biochemistry 1976; 15:4468 - 75; http://dx.doi.org/10.1021/bi00665a021; PMID: 823963
- Lattke H, Weser U. Yeast RNA-polymerase B: A zinc protein. FEBS Lett 1976; 65:288 - 92; http://dx.doi.org/10.1016/0014-5793(76)80131-7; PMID: 782912
- Wandzilak TM, Benson RW. Yeast RNA polymerase III: a zinc metalloenzyme. Biochem Biophys Res Commun 1976; 76:247 - 52; http://dx.doi.org/10.1016/0006-291X(77)90718-5; PMID: 800337
- Treich I, Riva M, Sentenac A. Zinc-binding subunits of yeast RNA polymerases. J Biol Chem 1991; 266:21971 - 6; PMID: 1939219
- Falchuk KH, Mazus B, Ber E, Ulpino-Lobb L, Vallee BL. Zinc deficiency and the Euglena gracilis chromatin: formation of an alpha-amanitin-resistant RNA polymerase II. Biochemistry 1985; 24:2576 - 80; http://dx.doi.org/10.1021/bi00331a027; PMID: 3925988
- Terhune MW, Sandstead HH. Decreased RNA polymerase activity in mammalian zinc deficiency. Science 1972; 177:68 - 9; http://dx.doi.org/10.1126/science.177.4043.68; PMID: 5041777
- Lyons TJ, Gasch AP, Gaither LA, Botstein D, Brown PO, Eide DJ. Genome-wide characterization of the Zap1p zinc-responsive regulon in yeast. Proc Natl Acad Sci U S A 2000; 97:7957 - 62; http://dx.doi.org/10.1073/pnas.97.14.7957; PMID: 10884426
- Zhao H, Butler E, Rodgers J, Spizzo T, Duesterhoeft S, Eide D. Regulation of zinc homeostasis in yeast by binding of the ZAP1 transcriptional activator to zinc-responsive promoter elements. J Biol Chem 1998; 273:28713 - 20; http://dx.doi.org/10.1074/jbc.273.44.28713; PMID: 9786867
- Zhao H, Eide DJ. Zap1p, a metalloregulatory protein involved in zinc-responsive transcriptional regulation in Saccharomyces cerevisiae. Mol Cell Biol 1997; 17:5044 - 52; PMID: 9271382
- Lee YJ, Lee CY, Grzechnik A, Gonzales-Zubiate F, Vashisht AA, Lee A, Wohlschlegel J, Chanfreau GF. RNA polymerase I stability couples cellular growth to metal availability. Mol Cell 2013; 51:105 - 15; http://dx.doi.org/10.1016/j.molcel.2013.05.005; PMID: 23747013
- Fath S, Milkereit P, Podtelejnikov AV, Bischler N, Schultz P, Bier M, Mann M, Tschochner H. Association of yeast RNA polymerase I with a nucleolar substructure active in rRNA synthesis and processing. J Cell Biol 2000; 149:575 - 90; http://dx.doi.org/10.1083/jcb.149.3.575; PMID: 10791972
- Nair U, Klionsky DJ. Molecular mechanisms and regulation of specific and nonspecific autophagy pathways in yeast. J Biol Chem 2005; 280:41785 - 8; http://dx.doi.org/10.1074/jbc.R500016200; PMID: 16230342
- Kvam E, Goldfarb DS. Nucleus-vacuole junctions and piecemeal microautophagy of the nucleus in S. cerevisiae. Autophagy 2007; 3:85 - 92; PMID: 17204844
- Richardson LA, Reed BJ, Charette JM, Freed EF, Fredrickson EK, Locke MN, Baserga SJ, Gardner RG. A Conserved Deubiquitinating Enzyme ControlsCell Growth by Regulating RNA Polymerase I Stability. Cell Rep 2012; 2:1 - 14; http://dx.doi.org/10.1016/j.celrep.2012.07.009; PMID: 22840390
- Amerik AY, Nowak J, Swaminathan S, Hochstrasser M. The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol Biol Cell 2000; 11:3365 - 80; http://dx.doi.org/10.1091/mbc.11.10.3365; PMID: 11029042
- Warner JR, Vilardell J, Sohn JH. Economics of ribosome biosynthesis. Cold Spring Harb Symp Quant Biol 2001; 66:567 - 74; http://dx.doi.org/10.1101/sqb.2001.66.567; PMID: 12762058
- Grummt I. Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev 2003; 17:1691 - 702; http://dx.doi.org/10.1101/gad.1098503R; PMID: 12865296
- Simm C, Lahner B, Salt D, LeFurgey A, Ingram P, Yandell B, Eide DJ. Saccharomyces cerevisiae vacuole in zinc storage and intracellular zinc distribution. Eukaryot Cell 2007; 6:1166 - 77; http://dx.doi.org/10.1128/EC.00077-07; PMID: 17526722
- Naryshkina T, Bruning A, Gadal O, Severinov K. Role of second-largest RNA polymerase I subunit Zn-binding domain in enzyme assembly. Eukaryot Cell 2003; 2:1046 - 52; http://dx.doi.org/10.1128/EC.2.5.1046-1052.2003; PMID: 14555487
- Drygin D, Rice WG, Grummt I. The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer. Annu Rev Pharmacol Toxicol 2010; 50:131 - 56; http://dx.doi.org/10.1146/annurev.pharmtox.010909.105844; PMID: 20055700