Abstract
Histone H2B ubiquitylation is involved in regulation of transcription and other DNA transacting processes. It is performed by a RING finger-containing ubiquitin ligase, Bre1, which is conserved from yeast to humans. Recent studies have implicated both transcriptional stimulatory and repressive functions of histone H2B ubiquitin ligase at the same or different genes as discussed here.
Histone proteins compactly package DNA to form chromatin inside the nucleus of eukaryotic cell, and undergo various covalent modifications to regulate DNA transacting processes such as transcription, replication, recombination, and repair.Citation1-Citation4 One important covalent modification is the monoubiquitylation of histone H2B at lysine 123 (K123) in yeast (K120 in mammals). Histone H2B ubiquitylation has been implicated to regulate transcriptional initiation as well as elongation.Citation5-Citation11 Further, it is involved in cell cycle arrest, gene silencing, splicing, replication stress, recombinational repair, meiosis, and apoptosis.Citation12-Citation20 Thus, histone H2B ubiquitylation is associated with various cellular functions, and its absence or misregulation is strongly correlated with a number of diseases including cancers.
A complex of Bre1, Rad6, and Lge1 is essential for histone H2B ubiquitylation in Saccharomyces cerevisiae.Citation2,Citation4,Citation10,Citation21-Citation24 Bre1 is an E3 ubiquitin ligase for the E2 ubiquitin-conjugating enzyme, Rad6. Bre1 is conserved from yeast to humans. RNF20 (RING finer protein 20; also known as BRE1A, hBRE1 or BRE1) and RNF40 (RING finger protein 40; also known as BRE1B) are the human orthologs of yeast Bre1, and form a heterodimer that functions as an E3 ubiquitin ligase for histone H2B monoubiquitylation.Citation12,Citation25 Importantly, a recent studyCitation26 has identified WAC as a functional partner of RNF20/RNF40 in regulation of histone H2B ubiquitylation. Depletion of either RNF20 or RNF40 impairs histone H2B ubiquitylation.Citation25 RAD6A (or HHR6A) and RAD6B (or HHR6B) are the human orthologs of yeast ubiquitin conjugase, Rad6.Citation25 Both RAD6A and RAD6B interact with RNF20/RNF40 for histone H2B ubiquitylation.Citation25 However, RAD6A has much higher abundance than RAD6B in human cells.Citation25 In fact, RAD6A constitutes the majority of cellular RAD6 protein pool, and thus, RAD6B siRNA does not cause significant change in the cellular level of histone H2B ubiquitylation, while depletion of RAD6A dramatically impairs histone H2B ubiquitylation.Citation25
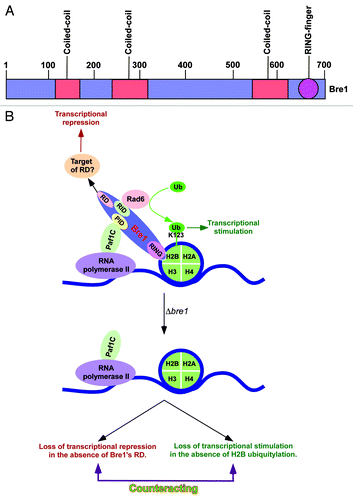
In yeast, Bre1 contains a C3HC4 Zn finger RING (really interesting new gene) near its C-terminus () that is essential for histone H2B ubiquitylation and subsequent histone H3 K4 methylation by COMPASS (Complex Proteins Associated with Set1), and histone H3 K79 methylation by Dot1 (1–4). The non-RING part of Bre1 contains several coiled-coil domains () and is involved in formation of homomeric complex through multiple intermolecular interactions.Citation21 Further, the non-RING domain of Bre1 interacts with ubiquitin conjugase, Rad6.Citation21 Such interaction is essential for targeted ubiquitylation of histone H2B.Citation21 Bre1 and Rad6 are recruited to the active chromatin by the interaction of Bre1 with Paf1 (polymerase II- associated factor 1; ). Further, Paf1 or Paf1-containing complex (Paf1C) is associated with transcribing RNA polymerase II. Thus, RNA polymerase II recruits Rad6 and Bre1 to the active chromatin for histone H2B ubiquitylation, and Paf1C plays an important role in such modification.
Figure 1. Transcriptional stimulatory and repressive functions of Bre1 at the same set of genes. (A) Schematic diagram showing different regions of Bre1 (1–700 amino acids). (B) Schematic diagram showing the stimulatory and repressive functions of Bre1 in transcription. Bre1 interacts with chromatin via its RING domain at the C-terminal.Citation21 Rad6 interacts with a domain within the first 210 amino acids at the N-terminal of Bre1,Citation21 and leads to targeted histone H2B ubiquitylation.Citation21 Paf1C interacts with Bre1Citation11,Citation21 via its non-RING domain to promote histone H2B ubiquitylation activity of Rad6-Bre1. Thus, bre1∆RING, ∆paf1, ∆rad6, and H2B-K123R mutant strains impair histone H2B ubiquitylation, RNA polymerase II association with active gene, and transcription.Citation5-Citation11,Citation21,Citation22 The non-RING part of Bre1 has a repression domain (RD) that lowers the association of RNA polymerase II with the active gene and hence transcription,Citation11 possibly via interaction with repressor(s) or by impairing the recruitment/activity of transcriptional stimulatory/elongation factor(s). The transcriptional stimulation is lost in the absence of histone H2B ubiquitylation in the bre1∆RING, ∆rad6, and H2B-K123R mutant strains. When the whole BRE1 is deleted, transcriptional repression is lost in the absence of the repression domain of Bre1, in addition to the impairment of transcriptional stimulation. These two opposing activities counteract, and hence the defect in transcription (or RNA polymerase II association with active gene) is not apparently observed in the ∆bre1 strain. PID, Paf1C interaction domain; RID, Rad6p interaction domain; Ub, ubiquitin.
Ubiquitylated-histone H2B is hydrolyzed to histone H2B by an ubiquitin protease, Ubp8, which is an integral component of the SAGA (Spt-Ada-Gcn5-acetyltransferase) complex.Citation27 Ubp8 is recruited to the promoter as a component of the SAGA complex.Citation27 It also associates with the coding sequence of active gene in a transcribing RNA polymerase II-dependent manner.Citation28 Thus, Ubp8 is involved in regulating the status of histone H2B ubiquitylation at the active gene. Histone H2B ubiquitylaion by Rad6-Bre1 and deubiquitylation by Ubp8 generates a dynamic histone ubiquitylation status at the active gene for efficient transcription.Citation22 Lge1 (Large 1) plays an important role in maintaining such a dynamic balance for histone H2B ubiquitylation and deubiquitylation.Citation22 Basically, Lge1 mediates the switch to histone H2B ubquitylation by attenuating Ubp8 recruitment and facilitating the association of Bre1 with chromatin.Citation22 Thus, the loss of Lge1 impairs histone H2B ubiquitylation.Citation22 Consistently, Madhani and colleagues have demonstrated that most of the phenotypes are shared by the ∆bre1 and ∆lge1 mutants.Citation23 However, it remains to be elucidated as to how Lge1 interacts with Bre1 to regulate Bre1 recruitment and histone H2B ubiquitylation in order to shed more insights into the Lge1 regulation of histone H2B ubiquitylation.
Previous studies have implicated histone H2B ubiquitylation in promoting transcriptional elongation.Citation5-Citation11 Such stimulation of transcriptional elongation is mediated via facilitated chromatin reassembly by ubiquitylation of histone H2B in the wake of transcribing RNA polymerase II.Citation7,Citation8 Accordingly, the absence of histone H2B ubiquitylation in the histone H2B-K123R point mutant strain impairs the association of RNA polymerase II with the GAL1 gene following transcriptional induction.Citation5,Citation11 Consistently, transcription of GAL1 is also impaired in the histone H2B K123R mutant strain.Citation5,Citation11 Thus, the depletion of the enzymes (such as Bre1 or Rad6) involved in histone H2B ubiquitylation is likely to decrease transcription as well as association of RNA polymerase II with GAL1. Indeed, the depletion of Rad6 is found to reduce the association of RNA polymerase II with GAL1 following transcriptional induction.Citation11 Consistently, GAL1 transcription is also decreased in the ∆rad6 strain in comparison to the wild type equivalent.Citation11 Surprisingly, complete depletion of Bre1 does not reduce GAL1 association of RNA polymerase II, and consequently, GAL1 transcription is not impaired in the ∆bre1 strain.Citation11 However, the deletion of Bre1’s RING domain (that is essential for histone H2B ubiquitylation; 21) significantly lowers the association of RNA polymerase II with GAL1, and hence transcription.Citation11 Therefore, the RING domain of Bre1 has a transcriptional stimulatory function, similar to the role of Rad6. However, the deletion of both RING and non-RING domains of Bre1 (or whole Bre1) does not decrease GAL1 association of RNA polymerase II (as well as transcription).Citation11 Thus, a non-RING domain of Bre1 appears to suppress the association of RNA polymerase II with GAL1 (and transcription), and counteracts the transcriptional stimulatory function of its RING domain (). Hence, GAL1 association of RNA polymerase II (as well as transcription) is not decreased in the ∆bre1 strain in comparison to the wild type equivalent.Citation11 Similar results are also found at several other genes such as GAL10, ADH1, and RPS5.Citation11 Taken together, these resultsCitation11 support both the stimulatory as well as repressive functions of Bre1 in regulation of RNA polymerase II association (and hence transcription) at the same set of genes via its RING and non-RING domains, respectively ().
The non-RING domain of Bre1 interacts with Paf1C to promote histone H2B ubiquitylation.Citation11,Citation21 Thus, the non-RING domain of Bre1 has a transcriptional stimulatory role via histone H2B ubiquitylation in addition to its transcriptional repressive function. However, it remains to be determined the region within Bre1’s non-RING domain that interacts with Paf1C. The results will define as to how Bre1 interacts with Paf1C, and hence, provide significant molecular insights into Paf1C regulation of histone H2B ubiquitylation and transcription. It is noteworthy to mention here that the association of RNA polymerase II with GAL1 in the ∆bre1 strain is not decreased, while the recruitment of RNA polymerase II-associated Paf1C is significantly impaired in the absence of Bre1.Citation11 These results support that Bre1 functions in targeting Paf1C to the active gene in addition to the role of RNA polymerase II in recruiting Paf1C. This is further substantiated by the observation of physical interaction between Bre1 and Paf1C.Citation21 Consistently, a recent study has also demonstrated the role of human BRE1/RNF20 in recruitment of human PAF1 complex (hPAF1C) for gene activation.Citation29
To determine the region within the non-RING domain of Bre1 involved in repressing RNA polymerase II association with active gene and transcription, Sen et al.Citation11 have analyzed the transcription regulatory functions of several Bre1 variants of different lengths. Such analysis reveals that the first ~200 amino acids at the N-terminal of Bre1 exhibit the repressive function in association of RNA polymerase II with active gene (and hence transcription). Further, a recent studyCitation21 demonstrated that the first 210 amino acids at the N-terminal of Bre1 interact with Rad6 for histone H2B ubiquitylation. Thus, the first ~200 amino acids in the non-RING domain of Bre1 have a transcriptional stimulatory role via histone H2B ubiquitylation in addition to its transcriptional repressive function ().
It is quite possible that the first ~200 amino acids region at the N-terminal of Bre1 interacts with certain repressor(s) to downregulate transcription. In fact, previous studiesCitation30-Citation33 have demonstrated the interaction of Bre1 with histone deacetylase that is involved in transcriptional repression. Thus, the first ~200 amino acids region within the non-RING domain of Bre1 may exhibit its transcriptional repressive function via histone deacetylation. Further, it may also be likely that the first ~200 amino acids region of Bre1 is essential for interaction with other repressor(s) to suppress transcription. Moreover, the ~200 amino acids region at the N-terminal of Bre1 may also repress transcription by impairing the recruitment or activity of certain transcriptional stimulatory/elongation factor(s). However, these possibilities need to be further investigated in order to shed mechanistic insights into the recently deciphered transcriptional repressive function of Bre1’s non-RING domain.
To determine the role of histone H2B ubiquitin ligase, Bre1, in global gene expression, Zhang et al.Citation34 performed DNA microarray analysis in conjunction with ubiquitin conjugase, Rad6, and H2B K123R point mutant. Such analysis reveals that transcription of many genes is altered in the absence of Rad6, thus indicating the role of Rad6-mediated histone H2B ubiquitylation in transcriptional regulation. However, all Rad6-regulated genes are not found to be overlapped with the genes whose transcription is altered in the histone H2B K123R mutant strain.Citation34 These results suggest an additional role of Rad6 in transcriptional regulation independently of histone H2B ubiquitylation. Surprisingly, transcription of many of these Rad6 and histone H2B ubiquitylation-regulated genes is not altered in the absence of Bre1.Citation34 It may be likely that transcription of these genes is dependent on the RING domain of Bre1, and the non-RING domain of Bre1 counteracts the transcriptional stimulatory function of its RING domain as observed in our recent studiesCitation11 at GAL1, GAL10, ADH1 and RPS5. Such possibility would not cause an apparent defect in transcription of these genes in the absence of Bre1. However, this needs to be tested by comparing genome-wide expression results of RING-deficient Bre1 with Bre1 null mutant. If the RING domain of Bre1 completely counteracts the repressive function of its non-RING domain at a set of genes, an alteration of transcription of those genes would be observed in the RING-deficient Bre1 mutant, but not in the Bre1 null mutant.
Interestingly, DNA microarray results in yeast reveal two distinct classes of genes that are regulated positively and negatively by Bre1.Citation34 Likewise, histone H2B ubiquitylation is involved in upregulation and downregulation of two distinct classes of genes.Citation34 Recent genome-wide studies by Pugh and colleaguesCitation8 have provided an important basis for such differential transcriptional regulation. Their results implicate that ubiquitylation of histone H2B is transcriptionally repressive at the promoter and activating at the coding sequence, while it favors nucleosomal assembly at both locations of gene. Histone H2B ubiquitylation-mediated nucleosomal assembly and/or stability prevents the formation of transcription machinery (and hence recruitment of RNA polymerase II) at the promoter, and therefore, lowers transcription.Citation8 The lowly transcribed genes have higher density of ubiquitylated-histone H2B at their promoter regions compared with coding regions.Citation8 Thus, lowly transcribed genes are negatively regulated by histone H2B ubuiquitylation via inhibition of the formation of transcription machinery at the promoter. On the other hand, histone H2B ubiquitylation at the transcribing regions of highly active genes promotes transcriptional elongation.Citation8 Ubiquitylated-histone H2B plays important role in nucleosomal reassembly in the wake of transcribing RNA polymerase II to promote transcription. The highly transcribed genes are enriched with ubiquitylated-histone H2B at their coding regions compared with promoter regions.Citation8 Thus, highly transcribed genes are regulated by histone H2B ubiquitylation at their coding regions. Taken together, histone H2B ubiquitylation regulates transcription differentially of highly and lowly transcribed genes based on the density of ubiquitylated-histone H2B at the promoter (repressive) or coding (activating) region.Citation8
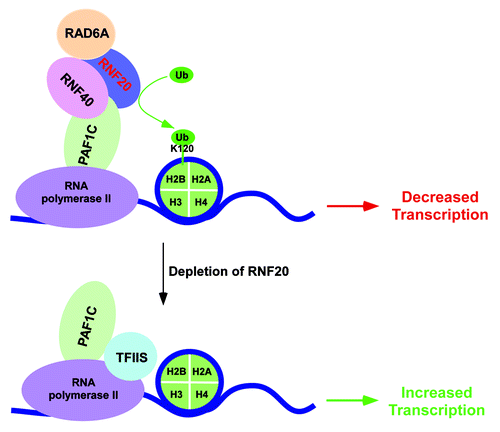
Like in yeast, human BRE1 or RNF20/RNF40 is involved in transcriptional stimulation and repression of two distinct sets of genes via different mechanisms.Citation12 The genes which are repressed by RNF20/RNF40 reside within compact chromatin.Citation12 Intriguingly, these genes are associated with significantly high levels of ubiquitylated-histone H2B as well as RNA polymerase II at their transcribed regions.Citation12 Other histone covalent modifications such as histone H3 K4 trimethylation and histone H3 K9 and 14 acetylation that are generally associated with gene activation are also found to be present at these RNF20-repressed genes.Citation12 Yet, these genes are expressed at low levels in the presence of RNF20, but upregulated in the absence of RNF20.Citation12 Possibly, compact chromatin structure encompassing these RNF20-repressed genes might hinder transcriptional elongation, resulting in paused/stalled RNA polymerase II at the transcribed regions. Such paused/stalled RNA polymerase II might require TFIIS to continue transcriptional elongation as TFIIS is involved in facilitating RNA polymerase II to pass through transcriptional pause/block on DNA by stimulating the endonucleolytic cleavage of nascent RNA. In support of this possibility, Shema et al.Citation35 have demonstrated that RNF20 prevents the targeting of TFIIS recruitment to the RNF20-repressed genes by interfering the interaction of TFIIS with hPAF1. TFIIS binds to both hPAF1 and RNA polymerase II.Citation12 However, the direct interaction of TFIIS with hPAF1 cooperatively enhances its association with RNA polymerase II. Further, hPAF1 mediates the association of RNF20/RNF40 with RNA polymerase II, and hence, plays an important role in histone H2B ubiquitylation, similar to yeast Paf1C. The enzymatic activity of RNA20-RNF40-hRAD6A has been implicated to interfere the binding of TFIIS with hPAF1C, possibly via histone H2B ubiquitylation (12, 35; ). Such interference decreases the association of TFIIS with RNF20-repressed genes, hence lowering transcription (12, 35; ). On the other hand, TFIIS association with RNF20-repressed genes is increased in the absence of RNF20, hence leading to transcriptional stimulation by unleashing RNA polymerase II (12, 35; ).
Figure 2. Schematic diagram showing the transcriptional repressive function of human BRE1 or RNF20/RNF40 at a set of genes. The enzymatic activity of RNF20/RNF40/RAD6A complex impairs the association of TFIIS with PAF1C (possibly via histone H2B ubiquitylation), hence lowering recruitment of TFIIS to the RNF20-repressed genes. Following depletion of RNF20, histone H2B ubiquitylation declines, and interaction between TFIIS and PAF1C is enhanced, which subsequently promotes recruitment of TFIIS to the chromatin to facilitate transcription.
While recent studies in human cells shed much light as to how RNF20 represses transcription,Citation12,Citation35 it remains obscure how RNF20 promotes transcription of a set of genes, but not others. Histone H2B ubiquitylation at the RNF20-dependent genes may be promoting recruitment of some positive elongation factor(s), alters chromatin structure or affect activity of some transcription factor(s) to facilitate transcription as proposed previously.Citation12 However, these possibilities remain to be investigated. Further, genome-wide analysis reveals that a large number of genes are not dependent on RNF20 for their transcription. Only a small subset of genes are regulated positively (~3%) or negatively (~3%) by RNF20.Citation12 It is not understood why a large number of genes are not regulated by RNF20, while there is a positive correlation between histone H2B ubiquitylation and transcription.
Although histone H2B ubiquitin ligase has been shown to regulate transcription of different sets of genes, it is yet unknown whether, similar to yeast Bre1, human BRE1 has transcriptional stimulatory and repressive functions at the same set of genes. Like in yeast, human BRE1 contains RING domain at its C-terminal, and coiled-coil-containing non-RING domain. Thus, similar to yeast Bre1, human BRE1 may have dual roles in transcriptional stimulation and repression at the same genes via its RING and non-RING domains, respectively. If transcriptional stimulatory function of BRE1’s RING domain is completely counteracted at a set of genes by its non-RING domain, no apparent defect in transcription of these genes would be observed in the absence of BRE1. Indeed, transcription of a set of genes in human cells is found to be unaffected in the absence of BRE1 or RNF20.Citation12 Thus, like in yeast, human BRE1 might be involved in transcriptional stimulation and repression at the same set of genes, which remains to be investigated.
In summary, recent studies in yeastCitation11 reveal that the RING domain of Bre1 has a stimulatory function in targeted histone H2B ubiquitylation for enhanced transcription, while Bre1’s non-RING domain has both the stimulatory and repressive roles in regulating the association of RNA polymerase II with active gene and hence transcription (). The stimulatory role of Bre1’s non-RING domain is mediated via its interaction with Rad6 and Paf1C, which facilitate histone H2B ubiquitylation (). The repressive function of the non-RING domain of Bre1 appears to be confined within its first ~200 amino acids. This function of Bre1 is likely to be crucial in slowing down transcribing RNA polymerase II for facilitating histone H2B ubiquitylation and subsequent chromatin reassembly for efficient transcription, since histone H2B ubiquitylation has been previously implicated in chromatin reassembly in the wake of transcribing RNA polymerase II.Citation7,Citation8 Thus, the transcriptional repressive role of Bre1 seems to be physiologically associated with its stimulatory function in histone H2B ubiquitylation, chromatin dynamics, and transcription. However, it remains to be investigated whether Bre1 ortholog performs similar functions in human cells.
Acknowledgments
The work in the Bhaumik laboratory was supported by a National Institutes of Health grant (1R15GM088798–01), a grant-in-aid (10GRNT4300059) from American Heart Association (Greater Midwest Affiliate), a Mallinckrodt Foundation grant, and Excellence in Academic Medicine (EAM) awards of SIU-School of Medicine. We apologize to the authors whose work could not be cited owing to space limitations.
Submitted
07/17/13
Revised
09/12/13
Accepted
09/26/13
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol 2007; 14:1008 - 16; http://dx.doi.org/10.1038/nsmb1337; PMID: 17984963
- Shukla A, Chaurasia P, Bhaumik SR. Histone methylation and ubiquitination with their cross-talk and roles in gene expression and stability. Cell Mol Life Sci 2009; 66:1419 - 33; http://dx.doi.org/10.1007/s00018-008-8605-1; PMID: 19370393
- Malik S, Bhaumik SR. Mixed lineage leukemia: histone H3 lysine 4 methyltransferases from yeast to human. FEBS J 2010; 277:1805 - 21; http://dx.doi.org/10.1111/j.1742-4658.2010.07607.x; PMID: 20236312
- Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem 2006; 75:243 - 69; http://dx.doi.org/10.1146/annurev.biochem.75.103004.142422; PMID: 16756492
- Shukla A, Bhaumik SR. H2B-K123 ubiquitination stimulates RNAPII elongation independent of H3-K4 methylation. Biochem Biophys Res Commun 2007; 359:214 - 20; http://dx.doi.org/10.1016/j.bbrc.2007.05.105; PMID: 17543890
- Xiao T, Kao CF, Krogan NJ, Sun ZW, Greenblatt JF, Osley MA, Strahl BD. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol Cell Biol 2005; 25:637 - 51; http://dx.doi.org/10.1128/MCB.25.2.637-651.2005; PMID: 15632065
- Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell 2008; 31:57 - 66; http://dx.doi.org/10.1016/j.molcel.2008.04.025; PMID: 18614047
- Batta K, Zhang Z, Yen K, Goffman DB, Pugh BF. Genome-wide function of H2B ubiquitylation in promoter and genic regions. Genes Dev 2011; 25:2254 - 65; http://dx.doi.org/10.1101/gad.177238.111; PMID: 22056671
- Tanny JC, Erdjument-Bromage H, Tempst P, Allis CD. Ubiquitylation of histone H2B controls RNA polymerase II transcription elongation independently of histone H3 methylation. Genes Dev 2007; 21:835 - 47; http://dx.doi.org/10.1101/gad.1516207; PMID: 17374714
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 2006; 125:703 - 17; http://dx.doi.org/10.1016/j.cell.2006.04.029; PMID: 16713563
- Sen R, Lahudkar S, Durairaj G, Bhaumik SR. Functional analysis of Bre1p, an E3 ligase for histone H2B ubiquitylation, in regulation of RNA polymerase II association with active genes and transcription in vivo.. J Biol Chem 2013; 288:9619 - 33; http://dx.doi.org/10.1074/jbc.M113.450403; PMID: 23417674
- Shiloh Y, Shema E, Moyal L, Oren M. RNF20-RNF40: A ubiquitin-driven link between gene expression and the DNA damage response. FEBS Lett 2011; 585:2795 - 802; http://dx.doi.org/10.1016/j.febslet.2011.07.034; PMID: 21827756
- Giannattasio M, Lazzaro F, Plevani P, Muzi-Falconi M. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J Biol Chem 2005; 280:9879 - 86; http://dx.doi.org/10.1074/jbc.M414453200; PMID: 15632126
- Zhang Y. Transcriptional regulation by histone ubiquitination and deubiquitination. Genes Dev 2003; 17:2733 - 40; http://dx.doi.org/10.1101/gad.1156403; PMID: 14630937
- Chernikova SB, Razorenova OV, Higgins JP, Sishc BJ, Nicolau M, Dorth JA, Chernikova DA, Kwok S, Brooks JD, Bailey SM, et al. Deficiency in mammalian histone H2B ubiquitin ligase Bre1 (Rnf20/Rnf40) leads to replication stress and chromosomal instability. Cancer Res 2012; 72:2111 - 9; http://dx.doi.org/10.1158/0008-5472.CAN-11-2209; PMID: 22354749
- Chernikova SB, Dorth JA, Razorenova OV, Game JC, Brown JM. Deficiency in Bre1 impairs homologous recombination repair and cell cycle checkpoint response to radiation damage in mammalian cells. Radiat Res 2010; 174:558 - 65; http://dx.doi.org/10.1667/RR2184.1; PMID: 20738173
- Walter D, Matter A, Fahrenkrog B. Bre1p-mediated histone H2B ubiquitylation regulates apoptosis in Saccharomyces cerevisiae. J Cell Sci 2010; 123:1931 - 9; http://dx.doi.org/10.1242/jcs.065938; PMID: 20460436
- Game JC, Chernikova SB. The role of RAD6 in recombinational repair, checkpoints and meiosis via histone modification. DNA Repair (Amst) 2009; 8:470 - 82; http://dx.doi.org/10.1016/j.dnarep.2009.01.007; PMID: 19230796
- Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 2002; 418:104 - 8; http://dx.doi.org/10.1038/nature00883; PMID: 12077605
- Zhang Z, Jones A, Joo HY, Zhou D, Cao Y, Chen S, Erdjument-Bromage H, Renfrow M, He H, Tempst P, et al. USP49 deubiquitinates histone H2B and regulates cotranscriptional pre-mRNA splicing. Genes Dev 2013; 27:1581 - 95; http://dx.doi.org/10.1101/gad.211037.112; PMID: 23824326
- Kim J, Roeder RG. Direct Bre1-Paf1 complex interactions and RING finger-independent Bre1-Rad6 interactions mediate histone H2B ubiquitylation in yeast. J Biol Chem 2009; 284:20582 - 92; http://dx.doi.org/10.1074/jbc.M109.017442; PMID: 19531475
- Song YH, Ahn SH. A Bre1-associated protein, large 1 (Lge1), promotes H2B ubiquitylation during the early stages of transcription elongation. J Biol Chem 2010; 285:2361 - 7; http://dx.doi.org/10.1074/jbc.M109.039255; PMID: 19923226
- Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, Madhani HD. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol Cell 2003; 11:261 - 6; http://dx.doi.org/10.1016/S1097-2765(02)00826-2; PMID: 12535538
- Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA, Dean K, Golshani A, Zhang Y, Greenblatt JF, et al. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol Cell 2003; 11:267 - 74; http://dx.doi.org/10.1016/S1097-2765(02)00802-X; PMID: 12535539
- Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell 2009; 137:459 - 71; http://dx.doi.org/10.1016/j.cell.2009.02.027; PMID: 19410543
- Zhang F, Yu X. WAC, a functional partner of RNF20/40, regulates histone H2B ubiquitination and gene transcription. Mol Cell 2011; 41:384 - 97; http://dx.doi.org/10.1016/j.molcel.2011.01.024; PMID: 21329877
- Shukla A, Stanojevic N, Duan Z, Sen P, Bhaumik SR. Ubp8p, a histone deubiquitinase whose association with SAGA is mediated by Sgf11p, differentially regulates lysine 4 methylation of histone H3 in vivo. Mol Cell Biol 2006; 26:3339 - 52; http://dx.doi.org/10.1128/MCB.26.9.3339-3352.2006; PMID: 16611979
- Wyce A, Xiao T, Whelan KA, Kosman C, Walter W, Eick D, Hughes TR, Krogan NJ, Strahl BD, Berger SL. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Mol Cell 2007; 27:275 - 88; http://dx.doi.org/10.1016/j.molcel.2007.01.035; PMID: 17643376
- Fonseca GJ, Cohen MJ, Nichols AC, Barrett JW, Mymryk JS. Viral retasking of hBre1/RNF20 to recruit hPaf1 for transcriptional activation. PLoS Pathog 2013; 9:e1003411; http://dx.doi.org/10.1371/journal.ppat.1003411; PMID: 23785282
- Pan X, Ye P, Yuan DS, Wang X, Bader JS, Boeke JD. A DNA integrity network in the yeast Saccharomyces cerevisiae.. Cell 2006; 124:1069 - 81; http://dx.doi.org/10.1016/j.cell.2005.12.036; PMID: 16487579
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. The genetic landscape of a cell. Science 2010; 327:425 - 31; http://dx.doi.org/10.1126/science.1180823; PMID: 20093466
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 2007; 446:806 - 10; http://dx.doi.org/10.1038/nature05649; PMID: 17314980
- Lin YY, Qi Y, Lu JY, Pan X, Yuan DS, Zhao Y, Bader JS, Boeke JD. A comprehensive synthetic genetic interaction network governing yeast histone acetylation and deacetylation. Genes Dev 2008; 22:2062 - 74; http://dx.doi.org/10.1101/gad.1679508; PMID: 18676811
- Zhang X, Kolaczkowska A, Devaux F, Panwar SL, Hallstrom TC, Jacq C, Moye-Rowley WS. Transcriptional regulation by Lge1p requires a function independent of its role in histone H2B ubiquitination. J Biol Chem 2005; 280:2759 - 70; http://dx.doi.org/10.1074/jbc.M408333200; PMID: 15528187
- Shema E, Kim J, Roeder RG, Oren M. RNF20 inhibits TFIIS-facilitated transcriptional elongation to suppress pro-oncogenic gene expression. Mol Cell 2011; 42:477 - 88; http://dx.doi.org/10.1016/j.molcel.2011.03.011; PMID: 21596312