Abstract
During transcription of protein coding genes by RNA Polymerase II the mRNA is processed and packaged into an mRNP. Among the proteins binding cotranscriptionally to the mRNP are mRNA export factors. One of the protein complexes thus coupling transcription to mRNA export is the TREX complex. However, despite the fact that TREX was identified and characterized about a decade ago, it had remained enigmatic how TREX is recruited to genes. The conserved Prp19 complex (Prp19C) has long been known for its function in splicing. We recently identified Prp19C to be essential for a second step in gene expression namely TREX occupancy at transcribed genes, answering this long-standing question but also raising new ones.
Keywords: :
Cotranscriptional Biogenesis of mRNPs
During transcription of protein-coding genes by RNA polymerase II (RNAP II) several mRNA-processing factors and mRNA-binding proteins associate with the nascent pre-mRNA to produce a mature messenger ribonucleoprotein particle (mRNP). The RNA itself is processed (addition of a 7-methyl-guanosine cap at the 5′ end, splicing, 3′ end cleavage and polyadenylation) by successively associating factors. A key element allowing the tight coupling of RNA processing events with transcription is the C-terminal domain (CTD) of Rpb1, the largest subunit of RNAP II. The CTD serves as a recruiting platform for the different processing factors and consists of repeats of the heptapeptide YSPTSPS that are differentially phosphorylated during the transcription cycle (reviewed inCitation1). The CTD is phosphorylated on serine 5 (S5) during transcription initiation, which recruits the capping enzyme. During transcription elongation S5 phosphorylation decreases and S2 phosphorylation increases. An S2 phosphorylated CTD is needed for recruitment of splicing, termination and polyadenylation factors to the transcription machinery. In addition, several mRNA-binding proteins, e.g., the SR-proteins Gbp2, Hrb1 and Npl3, are recruited to the transcription machinery and are transferred to the mRNA.Citation2,Citation3 After 3′ end processing (cleavage and polyadenylation) the mRNP is released from the site of transcription (reviewed in refs. Citation4,Citation5). Quality control mechanisms, which are still poorly understood, ensure that only mRNPs with a fully processed mRNA and a correct set of associated proteins are transported to the cytoplasm for translation.Citation6-Citation10
The TREX Complex Functions in Transcription, mRNP Formation and DNA Repair
In addition to mRNP processing factors, mRNA export factors bind cotranscriptionally to the mRNP. A key player coupling transcription elongation to mRNA export is the conserved TREX complex.Citation6-Citation8,Citation11 TREX consists of the THO subcomplex, comprised of Tho2, Hpr1, Mft1 and Thp2, the RNA export factors Sub2 and Yra1, the SR-proteins Gpb2 and Hrb1 and Tex1 (). TREX functions in transcription elongation as well as mRNA export. THO is required for recruitment of Sub2 and Sub2 for recruitment of Yra1, which in turn interacts with the mRNA export receptor Mex67-Mtr2 ( and refs. Citation6-Citation8,Citation11). However, TREX links transcription elongation not only to mRNA export but also to 3′-end formation (refs. Citation6,Citation7 and references therein). In addition, THO mutants exhibit transcription-associated hyperrecombination and impaired transcription elongation, particularly for long, G+C-rich, or internal repeat-containing genes (ref. Citation7 and references therein). Interestingly, Hpr1 and Tho2 also function in nucleotide excision repair (NER).Citation12 Thus, TREX is important for mRNP biogenesis as well as genome maintenance. In contrast to yeast, TREX is recruited to the mRNA in a splicing dependent manner in higher eukaryotes.Citation13 However, its functions in gene expression are thought to be conserved.
Figure 1. mRNP formation and export to the cytoplasm. Several mRNA binding proteins decorate the nascent mRNA forming an mRNP. One of the protein complexes coupling transcription to export of the mRNP is the TREX complex. The Prp19 complex (Prp19C) is essential for the occupancy of TREX at transcribed genes. The C-terminus of the Prp19C subunit Syf1 is necessary for efficient recruitment of Prp19C and TREX. After cleavage and polyadenylation the mRNP is released from the site of transcription and remodeled to an export competent mRNP. The export receptor Mex67-Mtr2 is recruited to the mRNP via direct interaction between the TREX component Yra1 and the mRNA export receptor. In addition, Hpr1, Nab2, Npl3 and the THSC-complex are implicated in Mex67-Mtr2 recruitment to the mRNP. Mex67-Mtr2 then exports the mRNP through the nuclear pore complex. Most S. cerevisiae proteins depicted here are conserved in higher eukaryotes.
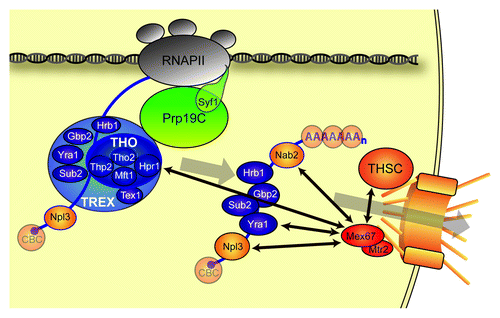
Recruitment of the mRNA Export Receptor Mex67-Mtr2 by Various Adaptor Proteins
The formation of export-competent mRNPs involves coordinated remodeling events, i.e., the dissociation and association of proteins (reviewed in ref. Citation4). The mRNP needs to recruit the conserved heterodimeric export receptor Mex67-Mtr2 in S. cerevisiae (Tap-p15 in higher eukaryotes) to be transported through the nuclear pore complex to the cytoplasm. This interaction is mediated by adaptor proteins. In addition to the TREX/Yra1-dependent recruitment of Mex67-Mtr2,Citation14 alternative export mechanisms have been suggested: First, Npl3, an heterogeneous nuclear ribonucleoprotein (hnRNP)- / SR-protein, has been implicated in recruiting Mex67 to the mRNA. Second, Hpr1 interacts directly with Mex67 suggesting a direct, i.e., Yra1-independent, recruitment mechanism (reviewed in refs. Citation6-Citation8). Furthermore, Nab2, another mRNA-binding protein, has been shown to be required for efficient export of bulk mRNA from the nucleus as well as poly(A) length control (ref. Citation5 and references therein).
Another complex required for mRNA export in S. cerevisiae is the Thp1-Sac3-Sus1-Cdc31 (THSC) complex which is—in contrast to the nucleoplasmic THO/TREX—located at the nuclear pore complex and tethers active genes to the nuclear rim (refs. Citation6,Citation7 and references therein). Its mutants exhibit similar hyperrecombination and impaired transcription phenotypes as mutants of the THO/TREX complex (ref. Citation7 and references therein).
Function of the Prp19 Complex in Splicing
The Prp19 complex (Prp19C) is a non-snRNA protein complex essential for splicing. It was primarily identified in S. cerevisiae as a complex containing Prp19 and at least eight other components.Citation15 Subsequently, the other components of the complex were identified, namely Ntc20, Snt309, Isy1, Syf2, Cwc2, Prp46, Clf1, Cef1 and Syf1.Citation16-Citation18 The Prp19 complex associates with the spliceosome during or after dissociation of the U4 snRNP, stabilizes the association of the U5 and U6 snRNPs in the activated spliceosomal B* complex, which catalyzes the first transesterification reaction of splicing, and remains bound through the second step of splicing.Citation19
Prp19, the core component of the complex, is essential and conserved across many eukaryotic species. The protein tetramerizes via its coiled-coil domain and forms a structural scaffold for the interaction with other components of the complex, such as Cef1, Snt309 and Cwc2.Citation20 The N-terminus contains a U-box domain which has E3 ubiquitin ligase activity in vitro. Mutations which demolish either Prp19-tetramer formation or the E3 ubiquitin ligase activity cause cell death.Citation20 Since Prp19 was first identified in a genetic screen for mutations conferring sensitivity to the DNA damaging reagent psoralen, it has long been thought to play a direct role in DNA repair.Citation21 However, in mutants of PRP19 the splicing efficiency of several transcripts encoding proteins involved in homologous recombination, DNA repair, cell cycle and chromosome segregation is impaired. This fact would sufficiently explain the defect of prp19 mutants in DNA damage repair, even though Prp19 might also function directly in DNA repair.
Another essential component of the Prp19 complex, Syf1, was originally identified as “synthetic lethal with cdc forty” in a screen for genes interacting with CDC40/PRP17.Citation22 Syf1 is highly conserved throughout evolution and consists mainly of 15 copies of the tetratricopeptide repeat (TPR) motif. Each motif consists of 34 amino acids with the highly degenerated consensus of W-L-G-Y-A-F-A-P. Regardless of its sequence diversity, the TPR motif has a conserved three-dimensional structure of a helix-turn-helix motif with adjacent TPR motifs packed into anti-parallel α helices. Syf1, as a multiple-TPR motif containing protein, is predicted to fold into a right-handed superhelical structure with a continuous helical groove suitable as scaffold for protein-protein interactions.
A Novel Function of Prp19C: The First Factor Identified to be Necessary for TREX Occupancy at Genes
Despite the fact that TREX was identified about a decade ago and has been extensively studied, it has remained enigmatic which factors are needed to recruit TREX to the gene. Thus, we aimed to answer this long-standing question. Interestingly, XAB2, the human homolog of Syf1, is a multifunctional protein important for gene expression, i.e., transcription and splicing, and maintenance of genome stability;Citation23,Citation24 functions that are very similar to the ones of the TREX complex. Thus, we assessed whether in S. cerevisiae Syf1 and the Prp19 complex could also have a function in transcription—maybe in conjunction with TREX. We showed that SYF1 genetically interacts with components of the TREX complex indicating indeed a functional overlap. Moreover, Syf1 also binds TREX in an RNA-independent manner. Syf1 and other proteins of Prp19C and thus most likely the whole Prp19 complex are recruited to the active transcription machinery of intron-containing but also intron-less genes, suggesting that Prp19C has a function in transcription independent of its function in splicing. Indeed, functional Syf1 is necessary for efficient transcription both in vivo and in vitro. Syf1 is needed for full processivity of RNAP II, i.e., its ability to travel the entire length of the gene—as is TREX. Interestingly, in a syf1 mutant lacking the C-terminus, Prp19C is not efficiently recruited to the transcription machinery and does not interact efficiently with RNAP II. Importantly, in this syf1 mutant recruitment of TREX is impaired, especially at the 3′-end of genes. We propose that Prp19C functions in transcription by stabilizing TREX at transcribed genes (). Taken together, we identified the first factor required for TREX occupancy at transcribed genes and thus a novel function of Prp19C in gene expression, namely transcription elongation (refs.Citation6,Citation25 and ).
Conservation of Prp19C and Its Subcomplexes
Since we isolated a syf1 mutant impaired in transcription but not splicing, these two functions of Prp19C are most likely independent. Thus, it is interesting to speculate that different subcomplexes exist that function in transcription vs. splicing. Based on genetic as well as biochemical experiments Prp19C consists of ten components in S. cerevisiae: Prp19, Syf1/Ntc90, Isy1/Ntc30, Clf1/Ntc77, Cef1/Ntc85, Syf2/Ntc31, Snt309/Ntc25, Ntc20, Prp46/Ntc50 and Cwc2/Ntc40.Citation17,Citation25,Citation26 However, even though one cannot exclude that the proteins identified in these pull-downs are a mixture of different subcomplexes, distinct subcomplexes have not been identified to date. Thus, in S. cerevisiae the Prp19C involved in transcription might be the same as the one involved in splicing.
In contrast, in human cells there are many more proteins, and there seem to be at least three different Prp19 complexes: The so called XAB2/Syf1 complex consists of XAB2/hSyf1, PRP19, ISY1, hAquarius, CCDC16 and PPIE.Citation24 The Prp19/CDC5L complex consists of PRP19, CDC5L, PRL1, SPF27 and the more loosely associated components CTNNBL1, HSP73 and AD002. The PRP19-related complexCitation27 consists of XAB2/hSyf1, ISY1, hAquarius/KIAA0560, CCDC12, PPIE/CypE, CRNKL1, RBM22, GCIP/SYF2, SKIP/PRP45, PPIL1, G10 and p29. All of these complexes were shown to have a function in splicingCitation24,Citation28 (see for composition and function of the different subcomplexes). However, only the XAB2 complex has been shown to function in transcription elongation and transcription coupled DNA repair (TCR),Citation24 which is similar to the function of TREX. Thus, even though a function of the other Prp19-complexes in transcription elongation and TCR has not been excluded, it is tempting to speculate, that in higher eukaryotes different Prp19 complexes have more specialized functions, i.e., all of them functioning in splicing but only the XAB2 complex in transcription elongation and TCR ().
Figure 2. Conservation of the Prp19 complex (Prp19C). In S. cerevisiae, one Prp19C has been identified that consists of ten subunits. In higher eukaryotes, e.g. H. sapiens, three different Prp19 complexes have been identified: The XAB2 complex, the PRP19/CDC5L complex and the PRP19-related complex. The proteins present in each complex are depicted in green. The position in the grid indicates the homology between S. cerevisiae and H. sapiens proteins. Cwc15 in S. cerevisiae (indicated by a star) has not been found in yeast Prp19C so far, but is homologous to human AD002 and was found to physically interact with yeast Clf1, a subunit of yeast Prp19C. Below each complex the known functions of each complex are given.
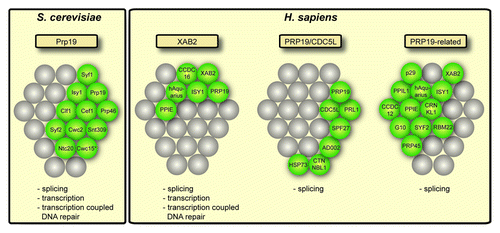
With the exception of Ntc20, all proteins of the S. cerevisiae Prp19C have human homologs and have been identified in one of the three PRP19 complexes (). The function of Prp19C in splicing is clearly conserved from yeast to higher eukaryotes. The function of Prp19C in recruitment of TREX to the mRNA might be also conserved. In contrast to yeast, the TREX complex is recruited to the mRNA during splicing in metazoan cells.Citation13 Interestingly, association of Prp19C with the spliceosome precedes recruitment of TREX.Citation29 Thus, Prp19C could also function in recruitment of TREX in higher cells. Which of the different Prp19 complexes might fulfill this function needs to be determined.
Open Questions Remain
Our work identified a novel function of Prp19C in transcription elongation by ensuring TREX occupancy at transcribed genes. However, a lot of open questions remain and new ones arise: First, Syf1 occupancy is reduced to about 50% in the syf1 mutant lacking the C-terminus but TREX occupancy at the 5′ end of genes is largely unaffected. Thus, Prp19C is most likely not required for the recruitment of TREX during transcription initiation but ensures TREX occupancy throughout the gene. It remains unknown how TREX is recruited to the 5′ end of genes. Is the emerging mRNA sufficient—in contrast to further downstream of the gene—or are other protein factors required to recruit TREX? The mRNA might be necessary for TREX recruitment since TREX binds to RNA and is not present at the promoter but recruited to the 5′-end of the open reading frame, i.e., at a time point when the newly synthesized mRNA emerges. Alternatively, but not mutually exclusive, other proteins such as Swt1, which interacts with TREX and functions in transcription as well as nuclear mRNP quality control,Citation30,Citation31 or an S2- or S2S5-(di)phosphorylated CTD might be important for recruitment of TREX. Interesting in this context, Sub2-independent mechanisms to recruit the TREX component Yra1 to the transcription machinery have been suggested: Yra1 interacts directly with the 3′ end processing factor Pcf11 consistent with a function of Yra1 in 3′ end processing.Citation32,Citation33 Second, it has been reported that Yra1 binds directly to the S2S5 di-phosphorylated CTD in vitro.Citation34 However, the functional relevance of these potentially alternative recruitment mechanisms of Yra1 and their potential interplay remain to be elucidated. In addition, it is not known whether the binding of Prp19C to TREX is direct or indirect. Thus, potential factor(s) required to recruit TREX to the gene at the promoter and/or any proteins that might “bridge” the interaction between Prp19C and TREX still need to be identified.
Second, since Prp19C stabilizes the interaction between TREX and RNAP II the question arises whether the interaction between Prp19C and RNAP II is direct and how and when Prp19C is recruited to the gene. Interesting in this context is the recent finding of a mechanism how splicing is enhanced by transcription or, more specifically, by the presence of the CTD of Rpb1, the largest subunit of RNAP II, in the mammalian system: Prp19C binds to another splicing factor, U2AF, and U2AF in turn binds to the S2-phosphorylated CTD.Citation35 Thus, also in S. cerevisiae Prp19C might be recruited to genes by binding to U2AF that interacts with elongating RNAP II. In addition, other mechanisms to recruit Prp19C to the transcription complex might exist. For example, Prp19C might bind directly to the S2- or S2/S5-diphosphorylated CTD. In any case, since recruitment of Prp19C to the transcription machinery is reduced to about 50% when the C-terminus of Syf1 is lacking, a second interaction interface between Prp19C and RNAP II still needs to be identified.
Third, TREX is dissociated from the gene approximately after cleavage of the nascent mRNA.Citation36 Thus, TREX could be released together with the mRNA it is bound to. Since Prp19C is required for continued TREX occupancy, it could play a pivotal role in dissociation of TREX. In addition, both dissociation of Prp19C and release of the mRNA could trigger efficient dissociation of TREX. However, a yet unknown mechanism could also be responsible for TREX dissociation by itself or in combination with Prp19C and the mRNA.
Fourth, the Prp19 subunit of Prp19C has an E3 ubiquitin ligase activity. Whereas this catalytic activity was shown to be essential for splicingCitation37 it is unclear whether it is also necessary for transcription elongation and TREX occupancy. If so, its substrate(s) need(s) to be identified.
Conclusion
In conclusion, we identified the first factor necessary for TREX occupancy at transcribed genes and uncovered a novel function of the Prp19 splicing complex in transcription elongation. However, as outlined above, many interesting questions remain to be answered.
| Abbreviations: | ||
| ChIP | = | chromatin immunoprecipitation |
| CTD | = | C-terminal domain |
| hnRNP | = | heterogenous nuclear ribonucleoprotein |
| mRNA | = | messenger RNA |
| mRNP | = | messenger ribonucleoprotein particles |
| Prp19C | = | Prp19 complex |
| RNAP II | = | RNA polymerase II |
| TPR | = | tetratricopeptide repeat |
Acknowledgments
We thank Robin Reed (Harvard Medical School) for critical reading of the manuscript. Grants from the German Research Foundation (DFG; SFB646) supported this research.
References
- Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev 2006; 20:2922 - 36; http://dx.doi.org/10.1101/gad.1477006; PMID: 17079683
- Hurt E, Luo MJ, Röther S, Reed R, Strässer K. Cotranscriptional recruitment of the serine-arginine-rich (SR)-like proteins Gbp2 and Hrb1 to nascent mRNA via the TREX complex. Proc Natl Acad Sci U S A 2004; 101:1858 - 62; http://dx.doi.org/10.1073/pnas.0308663100; PMID: 14769921
- Lei EP, Krebber H, Silver PA. Messenger RNAs are recruited for nuclear export during transcription. Genes Dev 2001; 15:1771 - 82; http://dx.doi.org/10.1101/gad.892401; PMID: 11459827
- Kelly SM, Corbett AH. Messenger RNA export from the nucleus: a series of molecular wardrobe changes. Traffic 2009; 10:1199 - 208; http://dx.doi.org/10.1111/j.1600-0854.2009.00944.x; PMID: 19552647
- Stewart M. Nuclear export of mRNA. Trends Biochem Sci 2010; 35:609 - 17; http://dx.doi.org/10.1016/j.tibs.2010.07.001; PMID: 20719516
- Molina-Navarro MM, Martinez-Jimenez CP, Rodriguez-Navarro S. Transcriptional Elongation and mRNA Export Are Coregulated Processes. Genetics Research International 2011; http://dx.doi.org/10.4061/2011/652461
- Rondón AG, Jimeno S, Aguilera A. The interface between transcription and mRNP export: from THO to THSC/TREX-2. Biochim Biophys Acta 2010; 1799:533 - 8; PMID: 20601280
- Rother S, Strasser K. mRNA export - an integrative component of gene expression. In Nuclear transport (ed R Kehlenbach), Landes Bioscience, Austin, TX 2009:101-17.
- Schmid M, Jensen TH. Quality control of mRNP in the nucleus. Chromosoma 2008; 117:419 - 29; http://dx.doi.org/10.1007/s00412-008-0166-4; PMID: 18563427
- Villa T, Rougemaille M, Libri D. Nuclear quality control of RNA polymerase II ribonucleoproteins in yeast: tilting the balance to shape the transcriptome. Biochim Biophys Acta 2008; 1779:524 - 31; PMID: 18644474
- Strässer K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 2002; 417:304 - 8; http://dx.doi.org/10.1038/nature746; PMID: 11979277
- González-Barrera S, Prado F, Verhage R, Brouwer J, Aguilera A. Defective nucleotide excision repair in yeast hpr1 and tho2 mutants. Nucleic Acids Res 2002; 30:2193 - 201; http://dx.doi.org/10.1093/nar/30.10.2193; PMID: 12000839
- Masuda S, Das R, Cheng H, Hurt E, Dorman N, Reed R. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev 2005; 19:1512 - 7; http://dx.doi.org/10.1101/gad.1302205; PMID: 15998806
- Strässer K, Hurt E. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J 2000; 19:410 - 20; http://dx.doi.org/10.1093/emboj/19.3.410; PMID: 10722314
- Tarn WY, Hsu CH, Huang KT, Chen HR, Kao HY, Lee KR, et al. Functional association of essential splicing factor(s) with PRP19 in a protein complex. EMBO J 1994; 13:2421 - 31; PMID: 8194532
- Chen CH, Tsai WY, Chen HR, Wang CH, Cheng SC. Identification and characterization of two novel components of the Prp19p-associated complex, Ntc30p and Ntc20p. J Biol Chem 2001; 276:488 - 94; http://dx.doi.org/10.1074/jbc.M006958200; PMID: 11018040
- Ohi MD, Gould KL. Characterization of interactions among the Cef1p-Prp19p-associated splicing complex. RNA 2002; 8:798 - 815; http://dx.doi.org/10.1017/S1355838202025050; PMID: 12088152
- Chen HR, Jan SP, Tsao TY, Sheu YJ, Banroques J, Cheng SC. Snt309p, a component of the Prp19p-associated complex that interacts with Prp19p and associates with the spliceosome simultaneously with or immediately after dissociation of U4 in the same manner as Prp19p. Mol Cell Biol 1998; 18:2196 - 204; PMID: 9528791
- Chan SP, Cheng SC. The Prp19-associated complex is required for specifying interactions of U5 and U6 with pre-mRNA during spliceosome activation. J Biol Chem 2005; 280:31190 - 9; http://dx.doi.org/10.1074/jbc.M505060200; PMID: 15994330
- Ohi MD, Vander Kooi CW, Rosenberg JA, Ren L, Hirsch JP, Chazin WJ, et al. Structural and functional analysis of essential pre-mRNA splicing factor Prp19p. Mol Cell Biol 2005; 25:451 - 60; http://dx.doi.org/10.1128/MCB.25.1.451-460.2005; PMID: 15601865
- Henriques JA, Moustacchi E. Isolation and characterization of pso mutants sensitive to photo-addition of psoralen derivatives in Saccharomyces cerevisiae. Genetics 1980; 95:273 - 88; PMID: 7009316
- Ben-Yehuda S, Dix I, Russell CS, McGarvey M, Beggs JD, Kupiec M. Genetic and physical interactions between factors involved in both cell cycle progression and pre-mRNA splicing in Saccharomyces cerevisiae. Genetics 2000; 156:1503 - 17; PMID: 11102353
- Nakatsu Y, Asahina H, Citterio E, Rademakers S, Vermeulen W, Kamiuchi S, et al. XAB2, a novel tetratricopeptide repeat protein involved in transcription-coupled DNA repair and transcription. J Biol Chem 2000; 275:34931 - 7; http://dx.doi.org/10.1074/jbc.M004936200; PMID: 10944529
- Kuraoka I, Ito S, Wada T, Hayashida M, Lee L, Saijo M, et al. Isolation of XAB2 complex involved in pre-mRNA splicing, transcription, and transcription-coupled repair. J Biol Chem 2008; 283:940 - 50; http://dx.doi.org/10.1074/jbc.M706647200; PMID: 17981804
- Chanarat S, Seizl M, Strässer K. The Prp19 complex is a novel transcription elongation factor required for TREX occupancy at transcribed genes. Genes Dev 2011; 25:1147 - 58; http://dx.doi.org/10.1101/gad.623411; PMID: 21576257
- Gahura O, Abrhámová K, Skruzný M, Valentová A, Munzarová V, Folk P, et al. Prp45 affects Prp22 partition in spliceosomal complexes and splicing efficiency of non-consensus substrates. J Cell Biochem 2009; 106:139 - 51; http://dx.doi.org/10.1002/jcb.21989; PMID: 19016306
- Deckert J, Hartmuth K, Boehringer D, Behzadnia N, Will CL, Kastner B, et al. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol Cell Biol 2006; 26:5528 - 43; http://dx.doi.org/10.1128/MCB.00582-06; PMID: 16809785
- Makarova OV, Makarov EM, Liu S, Vornlocher HP, Lührmann R. Protein 61K, encoded by a gene (PRPF31) linked to autosomal dominant retinitis pigmentosa, is required for U4/U6*U5 tri-snRNP formation and pre-mRNA splicing. EMBO J 2002; 21:1148 - 57; http://dx.doi.org/10.1093/emboj/21.5.1148; PMID: 11867543
- Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell 2009; 136:701 - 18; http://dx.doi.org/10.1016/j.cell.2009.02.009; PMID: 19239890
- Röther S, Clausing E, Kieser A, Strässer K. Swt1, a novel yeast protein, functions in transcription. J Biol Chem 2006; 281:36518 - 25; http://dx.doi.org/10.1074/jbc.M607510200; PMID: 17030511
- Skruzný M, Schneider C, Rácz A, Weng J, Tollervey D, Hurt E. An endoribonuclease functionally linked to perinuclear mRNP quality control associates with the nuclear pore complexes. PLoS Biol 2009; 7:e8; http://dx.doi.org/10.1371/journal.pbio.1000008; PMID: 19127978
- Johnson SA, Cubberley G, Bentley DL. Cotranscriptional recruitment of the mRNA export factor Yra1 by direct interaction with the 3′ end processing factor Pcf11. Mol Cell 2009; 33:215 - 26; http://dx.doi.org/10.1016/j.molcel.2008.12.007; PMID: 19110458
- Johnson SA, Kim H, Erickson B, Bentley DL. The export factor Yra1 modulates mRNA 3′ end processing. Nat Struct Mol Biol 2011; 18:1164 - 71; http://dx.doi.org/10.1038/nsmb.2126; PMID: 21947206
- MacKellar AL, Greenleaf AL. Cotranscriptional association of mRNA export factor Yra1 with C-terminal domain of RNA polymerase II. J Biol Chem 2011; 286:36385 - 95; http://dx.doi.org/10.1074/jbc.M111.268144; PMID: 21856751
- David CJ, Boyne AR, Millhouse SR, Manley JL. The RNA polymerase II C-terminal domain promotes splicing activation through recruitment of a U2AF65-Prp19 complex. Genes Dev 2011; 25:972 - 83; http://dx.doi.org/10.1101/gad.2038011; PMID: 21536736
- Kim M, Ahn SH, Krogan NJ, Greenblatt JF, Buratowski S. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J 2004; 23:354 - 64; http://dx.doi.org/10.1038/sj.emboj.7600053; PMID: 14739930
- Song EJ, Werner SL, Neubauer J, Stegmeier F, Aspden J, Rio D, et al. The Prp19 complex and the Usp4Sart3 deubiquitinating enzyme control reversible ubiquitination at the spliceosome. Genes Dev 2010; 24:1434 - 47; http://dx.doi.org/10.1101/gad.1925010; PMID: 20595234