Abstract
Maintenance of cell fates is essential for the development and homeostasis of multicellular organisms and involves the preservation of the expression status of selector genes that control many target genes. Epigenetic marks have pivotal roles in the maintenance of gene expression status, as occurs with methylation on lysine 27 of histone H3 (H3K27me) for Hox gene regulation. In contrast, because the levels of histone acetylation decrease during the mitotic phase, acetylated histone has not been believed to contribute to the maintenance of cell fates. Because members of the bromodomain and extra terminal (BET) family bind to acetylated histones localized on mitotic chromosomes, it is possible that they may regulate the transcriptional status of genes throughout the cell cycle. In this commentary, we discuss the recent analyses of C. elegans BET family protein BET-1, which contributes to the maintenance of cell fates through the histone H2A variant HTZ-1/H2A.z. This mechanism represses transcription of selector genes in the genomic region where lysine 27 of histone H3 (H3K27) is demethylated by histone demethylase UTX-1. We discuss the possibility that BET-1 and HTZ-1 maintain the poised state of RNA polymerase II in the cell such that it is ready to respond to differentiation signals.
The Maintenance of Cell Fates Through Histone Acetylation
Cell fate is determined by a combination of selector genes that encode transcription factors, which in turn, regulate many target genes.Citation1 Preserving the expression patterns of these selector genes is needed to maintain cell fates. Methylation on lysine 27 of histone H3 and Polycomb proteins maintain the repression of Hox genes.Citation2 In contrast to histone methylation, it is believed that histone acetylation does not function as an epigenetic mark that transmits cellular memory throughout the cell cycle, because the level of histone acetylation decreases during the mitotic phase.Citation3 Most of the acetylated histone-binding proteins do not associate with the mitotic chromosomes.Citation3 Exceptionally, BET family proteins that have two bromodomains do associate with mitotic chromosomes,Citation4-Citation8 and therefore, they are expected to play a role in maintaining cell fates. BET family proteins are evolutionarily conserved from yeast to human.Citation6 Characteristic features of BET family proteins are two bromodomains that bind to acetylated histone and an extra terminal (ET) domain.Citation6 BET family proteins are divided into two subfamilies, the short subfamily and the long subfamily; members of the latter subfamily are found only in multicellular organisms (). We have previously shown that BET-1, a C. elegans BET family protein that binds to acetylated histone H4,Citation8 is involved in the maintenance of cell fate in multiple lineages, including ectodermal and mesodermal lineages.Citation8
Table 1. BET family proteins
There has been recent and substantial progress in the study of BET family proteins with respect to medical research, especially in the fields related to cancer and viruses. For example, a fusion gene that consists of the mammalian BET family protein Brd4 and NUT (nuclear protein in testis) cause NUT midline carcinomas.Citation9 Constitutive expression of Brd2, another mammalian BET protein, also causes leukemia.Citation10 An inhibitor of BET proteins that interferes with binding of the bromodomain to acetylated histone has been studied as a drug for cancer therapy.Citation11,Citation12 It has also been reported that some viruses utilize BET family proteins for their own transcriptional regulation.Citation13,Citation14
BET family proteins also have important roles during normal development. The loss of mouse Brd4 causes post-implantation lethality.Citation15 Mutations in Drosophila fs(l)h that encodes BET family proteins cause homeotic transformation.Citation16-Citation18 Interestingly, homeotic transformation is also observed in mutants of Polycomb genes that are required for the maintenance of cell fates.Citation2 Therefore, the phenotype of fs(l)h mutants can be explained by the defect in the maintenance of cell fates. In addition, the role of BET family proteins as oncogenes suggests their role in the maintenance of cell fates.
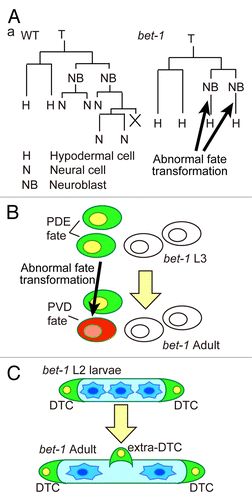
In C. elegans bet-1 mutants, abnormal cell fate transformation occurs in multiple cell lineagesCitation8 (). For example, transformation from the neural cell fate to the hypodermal cell fate was observed in the T-cell lineage (). Because bet-1 RNAi causes embryonic lethality at the morphogenesis stage, BET-1 may function as a component of the fundamental mechanism that is required for the maintenance of cell fates. Interestingly, in bet-1 mutants, cell fate transformation occurs even without cell division.Citation8 Therefore, BET-1 appears to be required for the maintenance of transcriptional status even after the final cell division. In addition, cells in bet-1 mutants transform to become closely related cells in terms of cell lineage. Transformation to the fate of sister or cousin cells is frequently observed.Citation8 Thus, BET-1 maintains the difference between closely related cells, including sister cells.
Figure 1. Phenotypes of bet-1 mutants in C. elegans. (A) The T-cell lineage in wild-type (WT) and bet-1 mutants. The posterior granddaughter cells show abnormal transformation of cell fate from neuroblasts to hypodermal cells. (B) Schematic drawing of bet-1 phenotype in the posterior lateral ganglia. In the wild-type V5.pa lineage, the PDE and PVD neurons express osm-6::gfp and dop-3::rfp, respectively. In bet-1 mutants, abnormal transformation from the PDE fate to the PVD fate was observed. (C) In the Z1/Z4 lineage, which produces the somatic gonad, wild-type animals produce two distal tip cells (DTCs) at the late L1 stage. In bet-1 mutants, abnormal transformation to the DTC fate occurs in the late larval stages, leading to the production of extra DTCs.
In addition to BET-1, the MYST family histone acetyltransferases MYS-1 and MYS-2 are required for the maintenance of cell fates.Citation8 BET-1 appears to bind to regions that are acetylated by MYS-1 and MYS-2. Their involvement indicates the importance of histone acetylation in the maintenance of cell fates. The positive charge of lysine in the histone tail is important for the interaction between histone and DNA in the nucleosome.Citation19 Histone acetylation on lysine loosens the interaction between histones and DNA. Therefore, it has been believed that histone acetylation correlates with transcriptional activation. However, in bet-1 mutants, ectopic expression of selector genes suggests that histone acetylation represses transcription through the activation of a BET-1-containing complex (see below).Citation20 Interestingly, genome-wide analyses in yeast suggest that each acetylation site appears to have a distinct role in transcriptional regulation, including transcriptional repression.Citation21 The mammalian BET family protein Brd4 is involved in repression of HPV (human papillomavirus) chromatin transcription.Citation14 Our results also suggests that BET-1 represses the transcription of ceh-22.Citation20 In addition, suppression of bet-1 phenotypes by RNAi of utx-1 that encodes H3K27 demethylase suggests that BET-1 and H3K27me has a similar effect in the maintenance of cell fates.Citation20 In addition, expression of egl-15 and sur-7 is upregulated in bet-1 mutants.Citation22 Thus, growing evidence indicates that histone acetylation also acts in transcriptional repression.
Transcriptional Repression by Histone Acetylation Through the Histone H2A Variant H2A.z in the Maintenance of Cell Fates
Because BET-1 itself does not have a catalytic domain, BET-1 is likely to function as a complex. In yeast, the BET family protein BDF1 is a part of the SWR1 complex, which is required for the deposition of the histone H2A variant HTZ1/H2A.z.Citation23 In C. elegans, htz-1/H2A.z functions in the same genetic pathway with bet-1 in the maintenance of cell fates.Citation20 The disruption of the SWR1 homolog SSL-1 causes the same phenotype that occurs in bet-1 mutants. Disruption of HTZ-1 in the mys-1 background also causes a defect in the maintenance of cell fates.Citation20 In addition, BET-1 regulates the subnuclear localization of HTZ-1.Citation20 Thus, in the maintenance of cell fates, BET-1 appears to function through the deposition of HTZ-1. HTZ-1 localizes on the transcription start site of the selector gene ceh-22, which is negatively regulated by HTZ-1 in the somatic gonad lineage, suggesting that BET-1-dependent HTZ-1 deposition directly regulates the transcription of selector genes. Thus, HTZ-1 appears to repress the transcription of selector genes in the maintenance of cell fates.
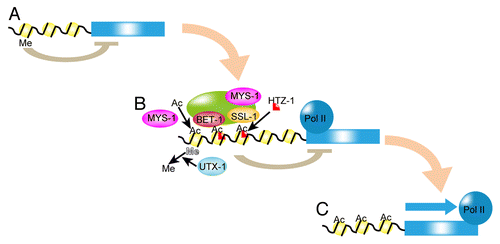
In C. elegans, genome-wide analysis of HTZ-1 indicates that the pattern of HTZ-1 occupancy on promoter regions is similar to that of RNA polymerase II.Citation24 There is, however, less of a correlation between HTZ-1 occupancy and transcriptional activity. Although H2A.z is implicated in transcriptional activation,Citation25-Citation27 there is no simple correlation between HTZ-1 localization and transcriptional activation.Citation24 One of the attractive hypotheses is that HTZ-1 regulates the pausing of RNA polymerase II on the transcription start site (). This hypothesis is consistent with the colocalization of HTZ-1 and RNA polymerase II and transcriptional repression by HTZ-1. In the poised state, RNA polymerase II is stalled near the transcription start site.Citation28 Therefore, the poised state is transcriptionally inactive but easily activated. Because the poised state is ready-to-go for transcriptional activation, in cells that are ready to respond to developmental signals, selector gene(s) may be in the poised state for quick response to developmental signals. Interestingly, in Drosophila, RNA polymerase II localizes on promoter regions of many transcriptionally inactive genes including developmental genes.Citation29 Because cells transform to the fates of closely related cells in terms of lineage in C. elegans bet-1 mutants, a transition to the poised state may also occur in sister or precursor cells of the cells that express selector genes. Maintenance of the poised state by HTZ-1 and BET-1 in the absence of a differentiation signal may prevent the over-production of specific cell types.
Figure 2. Working model for HTZ-1-dependent cell fate maintenance. (A) H3K27me silences the selector genes (blue box). Me, black line and yellow circle are methylation on H3K27, DNA, and histone, respectively. (B) UTX-1 demethylates H3K27me on selector gene loci. MYST histone acetyltransferases, including MYS-1 and MYS-2, acetylate the selector gene loci. Then, SSL-1 in the BET-1-containing complex deposits HTZ-1, which represses the transcription of selector genes. HTZ-1 may maintain the poised state of RNA polymerase II (Pol II). The transcription of selector gene(s) is ready to be activated. (C) When a cell receives a differentiation signal, repression by HTZ-1 is released and the gene is transcribed.
Histone acetylation recruits H2A.z to the selector gene loci through the action of BET-1 and SSL-1. However, how H2A.z maintains the resulting repression remains elusive. Transcriptional elongation by RNA polymerase II is promoted by phosphorylation at Ser2 on C-terminal domain (CTD).Citation28 Therefore, HTZ-1 might control the activity or recruitment of the P-TEFb complex that phosphorylates Ser2. Mammalian Brd4 associates with P-TEFb, suggesting that BET proteins may regulate C-terminal domain (CTD) phosphorylation at Ser2.Citation30
The regulation of the silencing mark H3K27me is also important for the maintenance of cell fate. Genome-wide analysis indicates that genomic localization of HTZ-1 and genomic localization of H3K27me are inversely correlated, suggesting that HTZ-1 and H3K27me occupy distinct regions of the genome.Citation20 In C. elegans, methylation on H3K27 is regulated by the histone methylase MES-2 and the histone demethylase UTX-1.Citation31 RNAi screening has identified utx-1 as a suppressor of the multiple bet-1 mutant phenotypes, including expression of the HTZ-1 target gene ceh-22.Citation20 This suggests that histone acetylation and H3K27me have similar effects in the maintenance of cell fates. Therefore, the relationship between BET-1, MYS-1, HTZ-1, and UTX-1 appears to be conserved for multiple selector genes. We also showed that, when utx-1 is disrupted, HTZ-1 localization to a target gene is decreased. Therefore, H3K27me appears to prevent the localization of HTZ-1 on the genome. In other words, HTZ-1 and BET-1 are recruited to the genomic region where H3K27 is demethylated by UTX-1. Thus, the HTZ-1-dependent mechanism appears to be an intermediate status between the H3K27me-related silenced state and transcriptional activation. Because the inverse correlation between H3K27me and H2A.z is also observed in differentiated mammalian cells,Citation32 this mechanism might be evolutionarily conserved.
Conclusion
Histone acetylation-dependent transcriptional repression of selector genes through H2A.z is important for the maintenance of cell fates. H2A.z may maintain the poised state of selector genes in the cell that are ready to respond to developmental signals. In contrast to transcriptional repression by H3K27me, the importance of transcriptional repression by histone acetylation and the poised state has been recognized only recently. Therefore, studying this mechanism should provide new insights into the transcriptional regulation that controls cell fates in animal development. C. elegans is advantageous for studying the maintenance of cell fate because of its invariant somatic cell lineage.Citation33 In addition, because the BET family protein Brd4 is known as a cancer-related gene, research in this field may also contribute to future cancer therapies.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by grants from the Japanese Ministry of Education, Culture, Sports, Science and Technology (Shibata Y and Nishiwaki K).
References
- Hobert O. Regulatory logic of neuronal diversity: terminal selector genes and selector motifs. Proc Natl Acad Sci U S A 2008; 105:20067 - 71; http://dx.doi.org/10.1073/pnas.0806070105; PMID: 19104055
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet 2004; 38:413 - 43; http://dx.doi.org/10.1146/annurev.genet.38.072902.091907; PMID: 15568982
- Kruhlak MJ, Hendzel MJ, Fischle W, Bertos NR, Hameed S, Yang XJ, Verdin E, Bazett-Jones DP. Regulation of global acetylation in mitosis through loss of histone acetyltransferases and deacetylases from chromatin. J Biol Chem 2001; 276:38307 - 19; PMID: 11479283
- Chua P, Roeder GS. Bdf1, a yeast chromosomal protein required for sporulation. Mol Cell Biol 1995; 15:3685 - 96; PMID: 7791775
- Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci U S A 2003; 100:8758 - 63; http://dx.doi.org/10.1073/pnas.1433065100; PMID: 12840145
- Florence B, Faller DV. You bet-cha: a novel family of transcriptional regulators. Front Biosci 2001; 6:D1008 - 18; http://dx.doi.org/10.2741/Florence; PMID: 11487468
- Kanno T, Kanno Y, Siegel RM, Jang MK, Lenardo MJ, Ozato K. Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Mol Cell 2004; 13:33 - 43; http://dx.doi.org/10.1016/S1097-2765(03)00482-9; PMID: 14731392
- Shibata Y, Takeshita H, Sasakawa N, Sawa H. Double bromodomain protein BET-1 and MYST HATs establish and maintain stable cell fates in C. elegans. Development 2010; 137:1045 - 53; http://dx.doi.org/10.1242/dev.042812; PMID: 20181741
- French CA, Ramirez CL, Kolmakova J, Hickman TT, Cameron MJ, Thyne ME, Kutok JL, Toretsky JA, Tadavarthy AK, Kees UR, et al. BRD-NUT oncoproteins: a family of closely related nuclear proteins that block epithelial differentiation and maintain the growth of carcinoma cells. Oncogene 2008; 27:2237 - 42; http://dx.doi.org/10.1038/sj.onc.1210852; PMID: 17934517
- Greenwald RJ, Tumang JR, Sinha A, Currier N, Cardiff RD, Rothstein TL, Faller DV, Denis GV. E mu-BRD2 transgenic mice develop B-cell lymphoma and leukemia. Blood 2004; 103:1475 - 84; http://dx.doi.org/10.1182/blood-2003-06-2116; PMID: 14563639
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011; 146:904 - 17; http://dx.doi.org/10.1016/j.cell.2011.08.017; PMID: 21889194
- Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, Robson SC, Chung CW, Hopf C, Savitski MM, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 2011; 478:529 - 33; http://dx.doi.org/10.1038/nature10509; PMID: 21964340
- Wu SY, Chiang CM. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem 2007; 282:13141 - 5; http://dx.doi.org/10.1074/jbc.R700001200; PMID: 17329240
- Wu SY, Lee AY, Hou SY, Kemper JK, Erdjument-Bromage H, Tempst P, Chiang CM. Brd4 links chromatin targeting to HPV transcriptional silencing. Genes Dev 2006; 20:2383 - 96; http://dx.doi.org/10.1101/gad.1448206; PMID: 16921027
- Houzelstein D, Bullock SL, Lynch DE, Grigorieva EF, Wilson VA, Beddington RS. Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Mol Cell Biol 2002; 22:3794 - 802; http://dx.doi.org/10.1128/MCB.22.11.3794-3802.2002; PMID: 11997514
- Gans M, Forquignon F, Masson M. The role of dosage of the region 7D1-7D5-6 of the X chromosome in the production of homeotic transformations in Drosophila melanogaster. Genetics 1980; 96:887 - 902; PMID: 6790337
- Huang DH, Dawid IB. The maternal-effect gene fsh is essential for the specification of the central region of the Drosophila embryo. New Biol 1990; 2:163 - 70; PMID: 1982070
- Chang YL, King B, Lin SC, Kennison JA, Huang DH. A double-bromodomain protein, FSH-S, activates the homeotic gene ultrabithorax through a critical promoter-proximal region. Mol Cell Biol 2007; 27:5486 - 98; http://dx.doi.org/10.1128/MCB.00692-07; PMID: 17526731
- Kimura A, Matsubara K, Horikoshi M. A decade of histone acetylation: marking eukaryotic chromosomes with specific codes. J Biochem 2005; 138:647 - 62; http://dx.doi.org/10.1093/jb/mvi184; PMID: 16428293
- Shibata Y, Sawa H, Nishiwaki K. HTZ-1/H2A.z and MYS-1/MYST HAT act redundantly to maintain cell fates in somatic gonadal cells through repression of ceh-22 in C. elegans. Development 2014; 141:209 - 18; http://dx.doi.org/10.1242/dev.090746; PMID: 24346701
- Kurdistani SK, Tavazoie S, Grunstein M. Mapping global histone acetylation patterns to gene expression. Cell 2004; 117:721 - 33; http://dx.doi.org/10.1016/j.cell.2004.05.023; PMID: 15186774
- Fisher K, Gee F, Wang S, Xue F, Knapp S, Philpott M, Wells C, Rodriguez M, Snoek LB, Kammenga J, et al. Maintenance of muscle myosin levels in adult C. elegans requires both the double bromodomain protein BET-1 and sumoylation. Biol Open 2013; 2:1354 - 63; http://dx.doi.org/10.1242/bio.20136007; PMID: 24285704
- Kobor MS, Venkatasubrahmanyam S, Meneghini MD, Gin JW, Jennings JL, Link AJ, Madhani HD, Rine J. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol 2004; 2:E131; http://dx.doi.org/10.1371/journal.pbio.0020131; PMID: 15045029
- Whittle CM, McClinic KN, Ercan S, Zhang X, Green RD, Kelly WG, Lieb JD. The genomic distribution and function of histone variant HTZ-1 during C. elegans embryogenesis. PLoS Genet 2008; 4:e1000187; http://dx.doi.org/10.1371/journal.pgen.1000187; PMID: 18787694
- Petty EL, Collette KS, Cohen AJ, Snyder MJ, Csankovszki G. Restricting dosage compensation complex binding to the X chromosomes by H2A.Z/HTZ-1. PLoS Genet 2009; 5:e1000699; http://dx.doi.org/10.1371/journal.pgen.1000699; PMID: 19851459
- Updike DL, Mango SE. Temporal regulation of foregut development by HTZ-1/H2A.Z and PHA-4/FoxA. PLoS Genet 2006; 2:e161; http://dx.doi.org/10.1371/journal.pgen.0020161; PMID: 17009877
- Meneghini MD, Wu M, Madhani HD. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 2003; 112:725 - 36; http://dx.doi.org/10.1016/S0092-8674(03)00123-5; PMID: 12628191
- Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu Rev Biochem 2012; 81:119 - 43; http://dx.doi.org/10.1146/annurev-biochem-052610-095910; PMID: 22404626
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet 2007; 39:1507 - 11; http://dx.doi.org/10.1038/ng.2007.21; PMID: 17994021
- Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell 2005; 19:535 - 45; http://dx.doi.org/10.1016/j.molcel.2005.06.029; PMID: 16109377
- Fisher K, Southall SM, Wilson JR, Poulin GB. Methylation and demethylation activities of a C. elegans MLL-like complex attenuate RAS signalling. Dev Biol 2010; 341:142 - 53; http://dx.doi.org/10.1016/j.ydbio.2010.02.023; PMID: 20188723
- Ku M, Jaffe JD, Koche RP, Rheinbay E, Endoh M, Koseki H, Carr SA, Bernstein BE. H2A.Z landscapes and dual modifications in pluripotent and multipotent stem cells underlie complex genome regulatory functions. Genome Biol 2012; 13:R85; http://dx.doi.org/10.1186/gb-2012-13-10-r85; PMID: 23034477
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol 1977; 56:110 - 56; http://dx.doi.org/10.1016/0012-1606(77)90158-0; PMID: 838129
