Figures & data
Figure 1. The bacterial flagellum. (a) Flagellar architecture and protein composition of Gram-negative (left) and Gram-positive (right) bacteria. (b) Domain architecture of Flagellin. The variable domains that are inserted into the conserved D1 domain (i.e. D1-N and D1-C in yellow and cyan, respectively) are shown in green. The conserved D0 that comprises N- and C-terminus of Flagellin is shown in red and blue, respectively. (c) Models of the Flagellin structures from S. typhimurium (pdb: 1UCU), Sphingomonas (based on pdb: 2ZBI), and Bacillus subtilis (this work). The color code is as in Figure (b). (d) Cartoon of the flagellar filament with the different Flagellins from Figure (c). The color code is as in Figure (b).
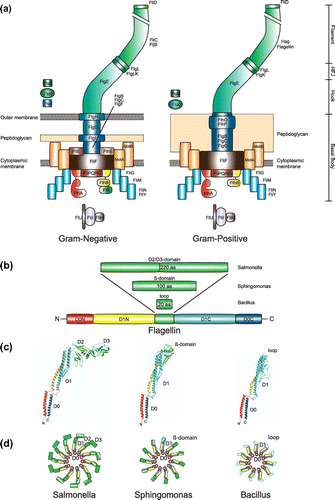
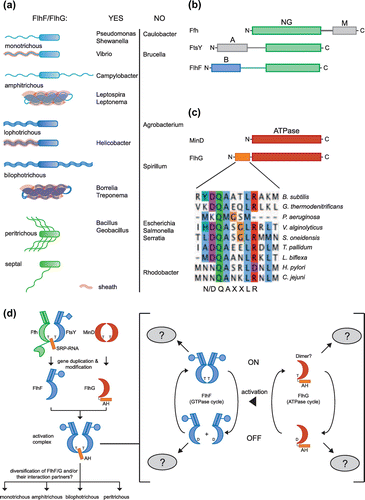
Figure 2. Bacterial flagellation patterns. (a) Polar flagellation patterns are colored in blue, and lateral patterns are marked in green. Sheathed flagella are shadowed in red. The listed organisms are grouped according to the occurrence and absence of FlhF and FlhG. (b) Domain architecture of SRP-GTPases. The conserved NG-domain is in green. The B-domain of FlhF is in blue. M- and A- domains of Ffh and FtsY, respectively, are in grey. (c) Domain architecture of MinD and FlhG. The N-terminus of FlhG contains the conserved ‘DQAxxLR’ motif. (d) Molecular evolution and diversification of the regulatory circuit of FlhF and FlhG. ‘T’ and ‘D’ stand for ATP/GTP and ADP/GDP, respectively. For further description see text.