Figures & data
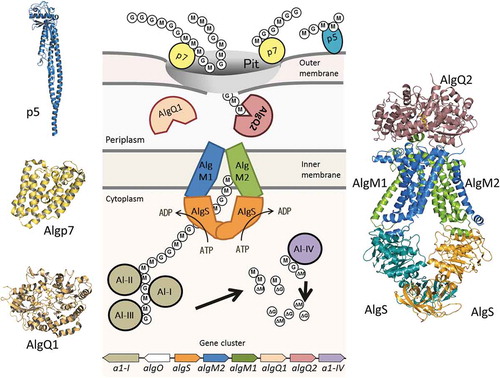
Figure 1. Alginate uptake in Sphingomonas sp. strain A1. An overall picture of alginate import and depolymerization systems. M and G represent the β-D-mannurorate and α-L-guluronate residues of alginate, respectively. AlgQ1 and AlgQ2 are solute-binding proteins that pass alginate to the ABC transporter. AlgM1, AlgM2, and AlgS form a heterotetramer (ABC transporter, AlgM1M2SS) and transport alginate across the inner membrane. A1-I to III are endo-type alginate lyases and A1-IV is an exo-type lyase. A1-I is divided to A1-II and A1-III. A series of actions by these lyases produce unsaturated monosaccharides (ΔM and ΔG), which are structurally the same. All of these proteins, together with a transcriptional regulator, AlgO, are coded by genes in the alginate-related gene cluster.
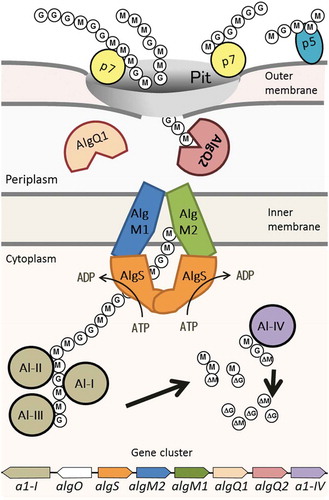
Figure 2. p5 and its derivatives.
(a) The correlation between amino acid deletions and alginate binding. (b) Tertiary structures of Salmonella flagellin and two p5 mutants. (c) Amino acid sequence alignment of N- and C-terminal regions of p5, p6, and Salmonella flagellin. Residues 20–40 and 353–363 of p5 are boxed.
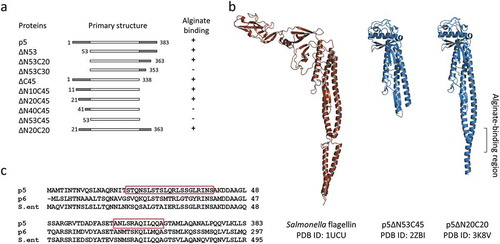
Figure 3. The structure of Algp7 (PDB ID; 5Y4C).
(a) The overall structure of Algp7 with bound copper ion (orange ball). (b) and (c) The surface structure of Algp7. The proposed alginate-binding site is circled (b), the copper ion-binding site is shown in the inset (c). Blue and red in the surface model represent positively and negatively charged amino acid residues, respectively. The orientation of (c) is the same with that of (a), but opposite to that of (b).
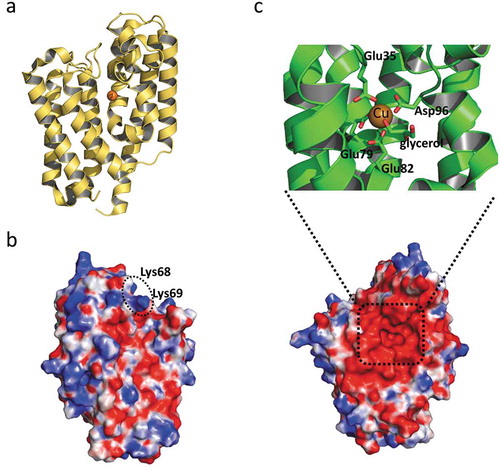
Figure 4. The structure of AlgQ1 and alginate oligosaccharide. (a) The structural difference between MMM and ΔGGG. Carbon atoms in MMM and ΔGGG are represented as yellow and green, respectively. (b) The overall structure of AlgQ1 in complex with MMM (PDB ID; 3VLU). (c) The alginate-binding site of AlgQ1 in complex with MMM (PDB ID; 3VLU). (d) The alginate-binding site of AlgQ1 in complex with ΔGGG (PDB ID; 3VLV). Hydrogen bonds formed between AlgQ1 and alginate are shown as dashed lines. The subsite number is shown in the parentheses in (c) and (d).
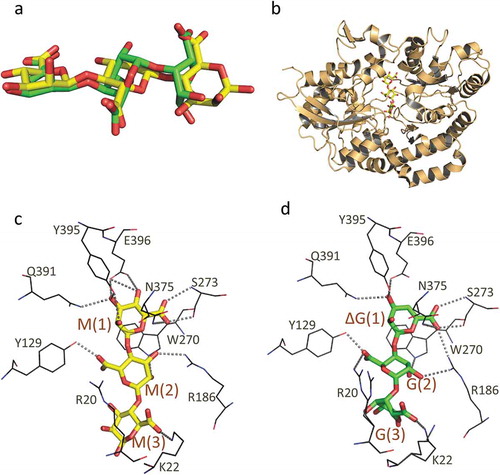
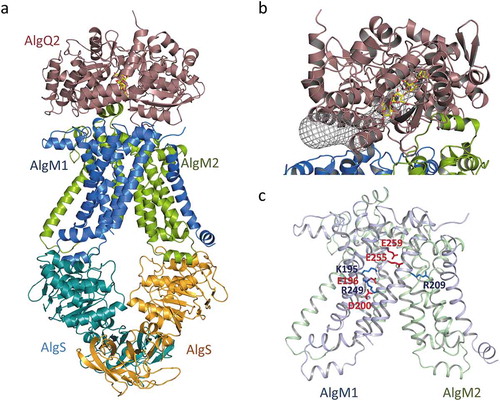
Figure 5. The crystal structure of the alginate transporter (PDB ID; 4TQU). (a) The overall structure of AlgM1M2SS in complex with AlgQ2. (b) The tunnel structure (grayed mesh) that continues to the alginate-binding site of AlgQ2. (c) Charged amino acid residues in transmembrane AlgM1M2. Blue and red represent basic and acidic residues, respectively.