Figures & data
Figure 1. Gene and domain organization of p53. TP53 gene and corresponding full-length protein (corresponds to human p53 major isoform α) is comprised of several functional domains. TAD1, transactivation domain 1; TAD2, transactivation domain 2; PRD, proline rich domain; DBD, DNA binding domain; NLS, nuclear localization signal; TETD, tetramerization domain; NES, nuclear export signal.
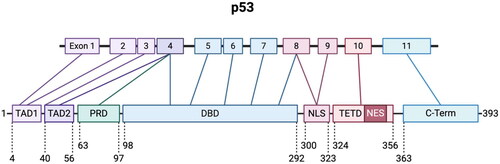
Figure 2. Canonical p53 stress response: cell cycle arrest and apoptosis. Growth arrest and apoptosis are well-understood p53 pathways in response to stress. Cell cycle arrest is mediated by transcriptional upregulation of p21, a cyclin-dependent kinase inhibitor that binds and inhibits CDK2. This causes decreased phosphorylation of the tumor suppressor RB, and RB is able to form the RB-E2F repressor complex to inhibit the upregulation of cell cycle genes. In order to induce apoptosis, p53 transcriptionally activates genes that include BAX, NOXA, and PUMA. BAX oligomerizes at the mitochondria to induce cytochrome c release to trigger the activation of caspase-9, which cleaves and activates caspase-3 and -7 to induce apoptosis. NOXA and PUMA inhibit the anti-apoptotic proteins BCL-2, BCL-XL, and MCL1, and this inhibition promotes BAK and BAX oligomerization at the mitochondria.
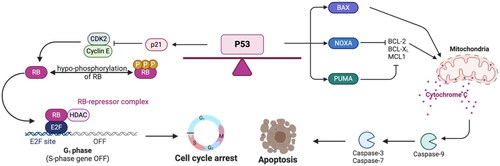
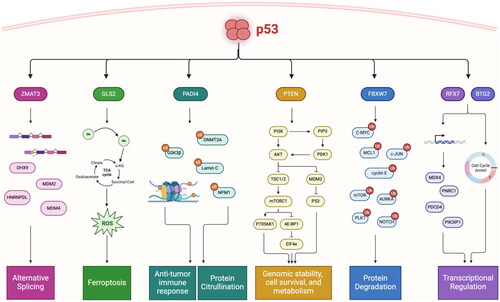
Figure 3. Chain of command: p53 transcriptionally activates “effectors” of tumor suppression that regulate distinct pathways. Critical target genes of p53 are mutated and inactivated in human cancers, have intrinsic tumor suppressive activity, and serve as master regulators of anti-tumor pathways. ZMAT3 regulates alternative splicing through binding to immature transcripts upstream of the 3’ splice site of critically important genes involved in tumor suppression and nonsense-mediated mRNA decay. GLS2 regulates ferroptosis by converting glutamine to glutamate to fuel the TCA cycle and generate reactive oxygen species (ROS) to trigger ferroptosis. PADI4 regulates immune activation through both transcriptional regulation and protein citrullination, and post translationally modifies key tumor suppressors to modulate their activity. PTEN collectively regulates genome stability, cell survival, and metabolism through the PI3K/AKT/mTOR pathway, as well as through inhibition of MDM2 to activate p53. FBXW7 is master regulator of protein degradation, and in particular controls the stability of key oncogenes in multiple cellular processes. RFX7 transcriptionally regulates downstream tumor suppressor genes, and BTG2 induces G1/S cell cycle arrest to protect against DNA-damage through its involvement in transcriptional regulation.