Figures & data
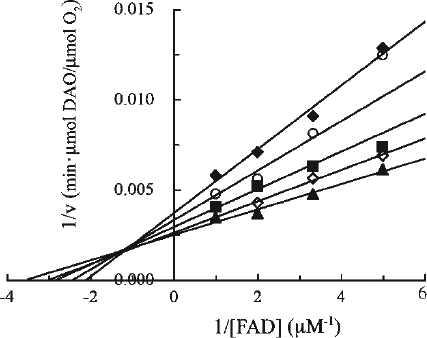
Figure 1. Kinetic analysis of the effect of CPZ on DAO. The reaction mixtures contained 5 μg of DAO (apoprotein), 50 mM D-alanine, FAD and CPZ as indicated, and 0.1 M sodium pyrophosphate (pH 7.3), in a total volume of 1.8 mL. The final CPZ concentrations were: none (open triangles), 200 μM (filled diamonds), 300 μM (open squares), and 400 μM (filled circles). Assays were carried out at 37°C. The data are representative of three experiments.
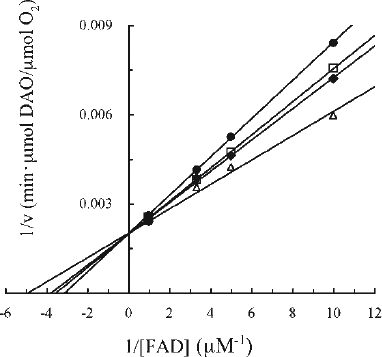
Figure 2. Elution profile of irradiated CPZ. The absorbances at 254 and 263 nm correspond to the λmax of CPZ and fraction #12, respectively. The effect of the fractions on DAO activity was examined at 25°C. The reaction mixtures contained 4 μg of DAO (apoprotein), 50 mM D-alanine, 0.3 μM FAD, 100 μL effluent and 0.1 M sodium pyrophosphate (pH 7.3), in a total volume of 1.8 mL. The ratio of DAO activity in the presence of each fraction was determined relative to the activity measured in the absence of effluent.
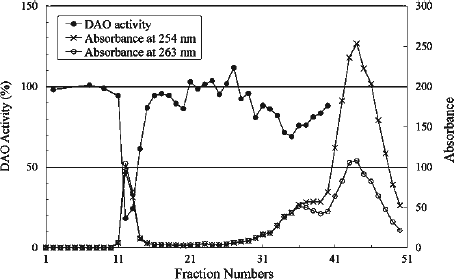
Figure 3. Absorption spectra of CPZ (20 μM), gel filtration fractions #12, 36 and 44. Fractions were diluted with H2O. The dilution factors for fractions #12, 36 and 44 were 1/100, 1/50 and 1/200, and the λmax for the fractions were 263, 257 and 254, respectively. Authentic CPZ has λmax at 254 nm.
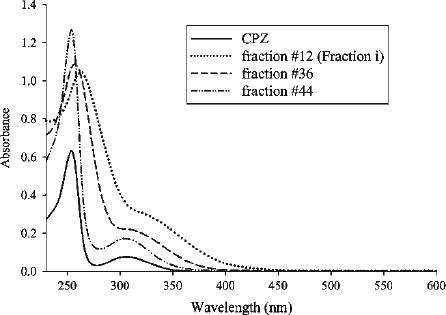
Figure 4. Examination for radicals. (a) EPR spectra of Fraction i in D2O (estimated to be 2 mM based on the measurement of dry weight) obtained without (top) and under (centre) UV irradiation, and EPR spectrum for 56 mM CPZ in D2O measured under UV irradiation (bottom). EPR conditions were: power, 20 mW; sweep width, 3358.4 ± 25 Gauss; modulation amplitude, 0.2 Gauss; time constant, 0.1 s; sweep time, 4 min; gain, 2 000 (top), 1 000 (centre), 500 (bottom). (b) Effect of radicals on DAO activity. The reaction mixtures contained 5 μg of DAO (apoprotein), 50 mM D-alanine, 0.3 μM FAD, 20 μL of Fraction i (estimated to be 2.9 μM based on the measurement of dry weight) and 1 mM ascorbate as indicated and 0.1 M sodium pyrophosphate (pH 7.3), in a total volume of 1.8 mL. Fraction i and ascorbate were preincubated for 3 min, then FAD and the apoprotein were added. After another 3-min incubation, D-alanine was added as substrate. Assays were carried out at 25°C. Data are the mean ± SD of three experiments. *p < 0.01, compared with control measured in the absence of Fraction i and ascorbate (one-way ANOVA with post-hoc Tukey's multiple comparison test).
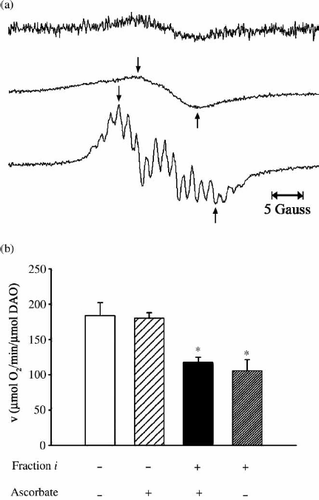
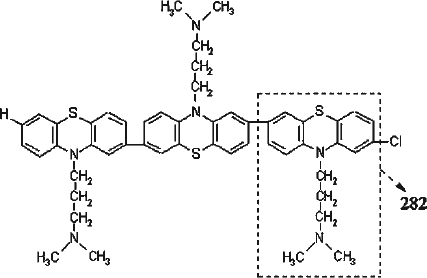
Figure 5. Line-broadening effect in 1H-NMR spectrum of Fraction i. The inset shows the structure of CPZ with the phenothiazine nucleus designated as A, the side chain carbon and methyl groups designated as α-γ and B, respectively. (Top) 1H spectrum (CD3OD at 400 MHz) of Fraction i. (Bottom) 1H spectrum (CD3OD at 400 MHz) of CPZ. The strong peaks at δ3.35 and δ4.9 in both spectra are due to CD2HOD and CD3OH, respectively.
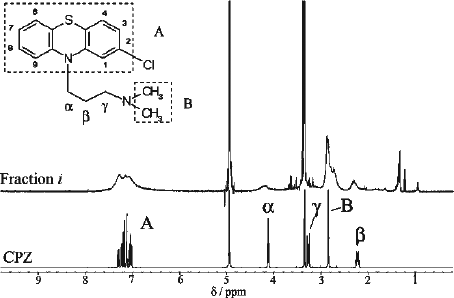
Figure 6. ESI (positive ionization mode) mass spectrum of Fraction i obtained by summing the area beneath the peak on TIC (see text).
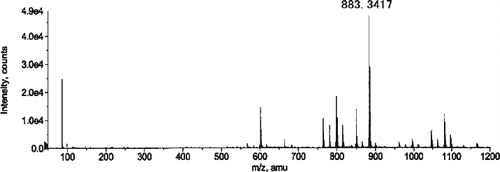
Table I. Summary Of Esi-tof-ms Analysis Of Cpz And Oligomers.
Figure 7. Kinetic analysis of the effect of Fraction i on DAO. The reaction mixtures were essentially the same as described in the legend to . Final Fraction i concentrations were: none (filled triangles), 0.92 μM (open diamonds), 1.5 μM (filled squares), 2.8 μM (open circles), and 3.7 μM (filled diamonds). Assays were carried out at 25°C. The data are representative of three experiments. Data points obtained at 1.5 μM and 2.8 μM of Fraction i in the presence of the lowest concentration of FAD were ignored.