Figures & data
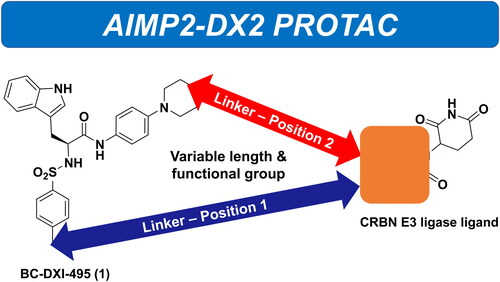
Figure 2. Chemical Structures of BC-DXI-495 (1) and putative positions to link the E3 ligase ligand.

Scheme 1. Synthesis of AIMP2-DX2 PROTACs 13–16, 18–23. Reagents and conditions: (i) 4-cyanobenzenesulfonyl chloride, TEA, THF:H2O, 0–25 °C, overnight; (ii) 4-morphlinoaniline, HATU, DIPEA, DMF, 25 °C, overnight; (iii) LiAlH4, THF, 0–25 °C, overnight; (iv) ADMP, DCM, DMAP, 25 °C overnight; (v) (+)-Sodium L-ascorbate, copper(II) sulphate pentahydrate, DMSO, 25 °C, overnight.

Scheme 2. Synthesis of AIMP2-DX2 PROTACs 25, 26, 28–31, and 33–34. Reagents and conditions: (i) KOH, 2-propanol, reflux, 18 h; (ii) HATU, DIPEA, DMF, 25 °C, overnight; (iii) HATU, DIPEA, DMF, 25 °C, overnight; (iv) HATU, DIPEA, DMF, 25 °C, overnight.

Scheme 3. Synthesis of AIMP2-DX2 PROTACs 36, 37, and 39–41. Reagents and conditions: (i), (ii) HATU, DIPEA, DMF, 25 °C, overnight.

Scheme 4. Synthesis of AIMP2-DX2 PROTACs 45–49. Reagents and conditions: (i) p-Toluenesulfonyl chloride, NEt3, THF:H2O (10:1 v/v mixture) 0–25 °C, overnight; (ii) HATU, DIPEA, DCM, 25 °C, 1 h; (iii) 4-(piperazin-1-yl) aniline, HATU, DIPEA, DMF, 60 °C, 4 h.

Scheme 5. Synthesis of AIMP2-DX2 PROTACs 51–54. Reagents and conditions: (i) 4-(piperazin-1-yl) aniline, K2CO3, MECN, reflux, overnight; (ii) compound 42, HATU, DIPEA, DCM, 25 °C, 1 h, overnight.

Table 1. Structures and the % inhibition AIMP2-DX2 values for PROTACs. 
Table 2. Structures and the % inhibition AIMP2-DX2 values for PROTACs with different linkers.
Table 3. IC50 value of compounds against AIMP2-DX2.
Figure 5. Compound 45 mediated induction of interaction between Siah1 and AIMP2-DX2. (A) Immunoprecipitation assay showing compound 45-dependent increased binding of two proteins. IP and WCL mean immunoprecipitation and whole cell lysates, respectively. Actin was used as a loading control. (B) In vitro pull-down assay confirming compound 45-mediated direct effect on binding of two proteins. GST proteins were detected by Coomassie staining. EV means empty vector.

Figure 6. Ubiquitination-mediated degradation of AIMP2-DX2 via compound 45. (A) Compound 45-mediated alteration of AIMP2-DX2 protein and mRNA level. Cells treated with compound 45 were subjected to western blotting (WB) and RT-PCR (RT). (B) Determination of compound 45-dependent ubiquitination of AIMP2-DX2. The cells expressing strep-AIMP2-DX2 were treated with compound 45 and MG132 and subjected to ubiquitination assay. Ub: ubiquitin.

Figure 7. Dependency of compound 45-mediated anti-proliferative efficacy on level of both AIMP2-DX2 and Siah1. (A) Siah1-dependency of the suppressed cell viability via compound 45. Siah1-knockdowned cells via its specific si-RNA were treated with compound and subjected to MTT assay. The results were shown as a bar graph (left) and the level of proteins were checked by immunoblotting (right). (B) Compound 45-mediated cell viability in lung cancer cell lines with the different level of AIMP2-DX2 and Siah1. Cell viability was determined as above. N.D.: not determined. (A and B) The experiments were independently repeated three times.