Figures & data
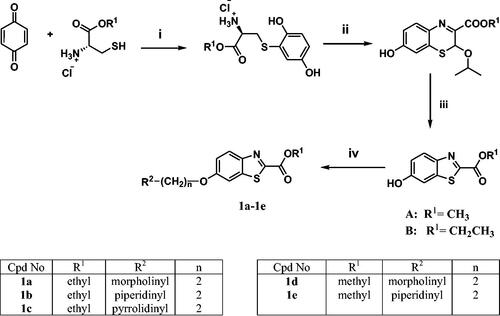
Scheme 1. Reagents and conditions: (i) MeOH, RT, overnight, (ii) K3Fe(CN)6, 4 M NaOH, aq. isopropanol, RT, overnight, (iii) 1 M HCl, EtOH, RT, overnight, (iv) 2 equiv. of chloro alkyl amine hydrochloride, 4 equiv. of K2CO3, KI, acetone, reflux, overnight.
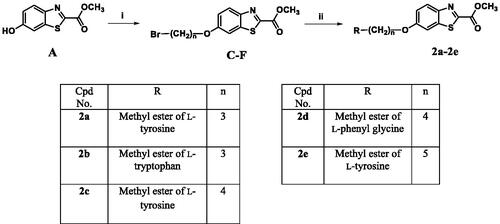
Scheme 2. Reagents and conditions: (i) 3 equiv. of α,ω-dibromoalkane, 4 equiv. of K2CO3, KI, acetone, reflux, overnight, (ii) 3 equiv. of the methyl ester of the aromatic amino acid, MeOH, reflux, overnight.
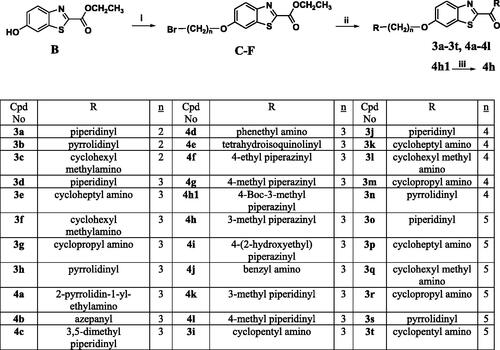
Scheme 3. Reagents and conditions: (i) 3 equiv. of α,ω-dibromoalkane, 4 equiv. of K2CO3, KI, acetone, reflux, overnight, (ii) 3 equiv. of appropriate amine, MeOH, reflux, overnight, (iii) TFA, DCM, 0 °C, RT, 2 h for Boc-protected derivative.
Scheme 4. Reagents and conditions: (i) 4 equiv. of glacial acetic acid, 4 equiv. of DMAP, 4 equiv. of EDC in DCM and DMF in ratio 1:1, RT, overnight.
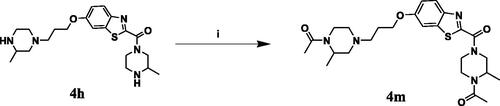
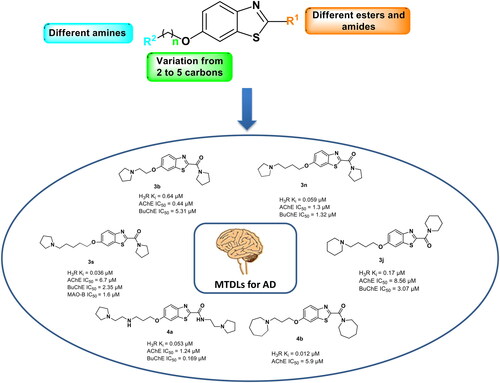
Table 1. Screening at H1R, H3R, H4R, and AChE by the first cluster of compounds (1a–1e).
Table 2. Inhibition of H1R, H3R, H4R and AChE by the 2nd cluster of compounds (2a–2e).
Table 3. Inhibition of H1R, H3R, H4R and AChE by 3rd cluster of compounds (3a–3t). 
Table 4. Inhibition of H3R, and AChE (compounds 4a–4m). 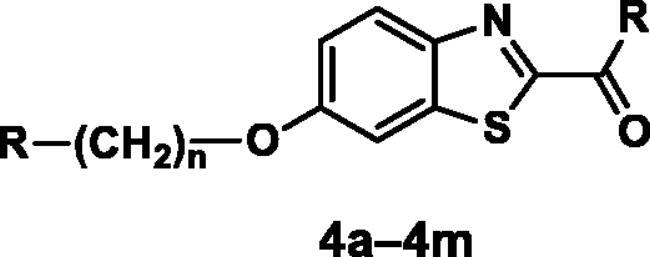
Figure 3. Binding mode of compound 3s within the homology model of H3R. 3s was docked into the active site using MOE. In the binding model, compound 3s formed three hydrogen bond interactions with Glu176, Cys88, and Arg351, a salt bridge with Glu176 and two CH–π interactions involving Trp341 and Phe163. Interactions are indicated by dashed lines and distances between the heavy atoms are given in Å.
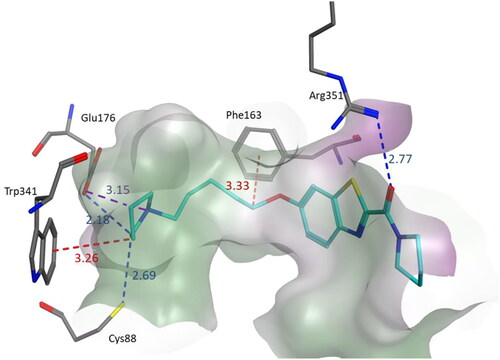
Table 5. Inhibition of selected compounds at BuChE and MAO-A and B enzymes.
Figure 4. Binding mode of compound 3b within the homology model of H3R. 3b was docked into the active site using MOE. In the binding model, compound 3b formed three hydrogen bond interactions with Glu176, a salt bridge with Glu176 and one hydrogen bond with Arg351. Dashed lines indicated the interactions and distances between the heavy atoms are given in Å.
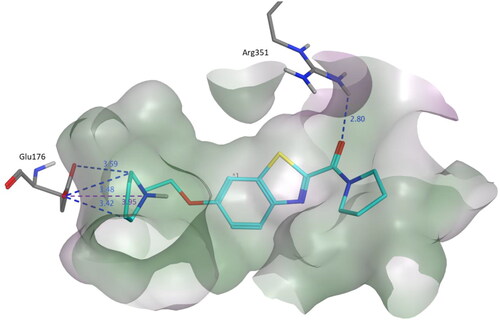
Figure 5. Binding mode of compound 3b within the AChE active site (PDB code: 4EY5). 3b was docked into the active site of AChE using MOE. In the binding model, compound 3b is anchored between amino acids Phe295, Tyr337, Glu202 and Trp86. It formed cation-pi interaction with Trp86. In addition, we found two CH–π interactions involving Tyr337 and two hydrogen bond interactions with Glu202 and Phe295. Dashed lines indicate the interactions and distances between the heavy atoms are given in Å.
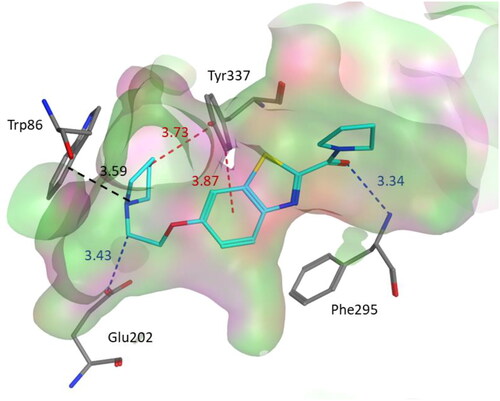
Figure 6. Binding mode of compound 4a within the BuChE active site (PDB code: 1POI). 4a was docked into the active site of BuChE using MOE. In the binding model, compound 4a is anchored between amino acids Phe329, Thr284, Ala277 and Glu197. It formed three hydrogen bond interactions involving Glu197, Ala277 and Thr284. In addition one CH–π interaction with Phe329. Dashed lines indicate the interactions and distances between the heavy atoms are given in Å.
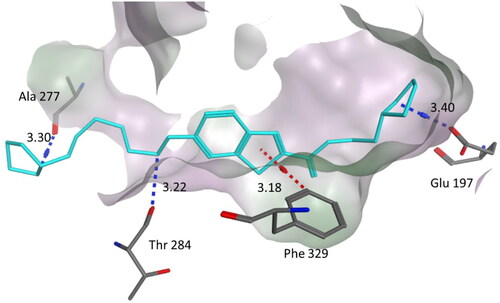
Table 6. Calculated physicochemical properties of compounds (3b, 3h, 3j, 3n, 3s, 3t, 4a, 4b)*.