Figures & data
Figure 1 Alignment of Cx50 and Cx32 protein sequences outlining the four transmembrane domains, M1, M2, M3, and M4 with a dashed box. Letters in red and blue text are sites that where cysteine substitutions were created for pore-lining studies of Cx32 and Cx50 respectively. Helical net plots are shown below for transmembrane domains of each connexin. Non-functional sites are highlighted with red circles and blue circles (Cx32 and Cx50, respectively) and indicate sites where cysteine substitution rendered the channels nonfunctional during scanning cysteine mutagenesis. Pore-lining residues in Cx32 are highlighted in light blue (Skerrett et al. Citation2002). Sequences were aligned using the ALIGN Optimal Global Alignment function (Biology Workbench, San Diego Super Computer Center).
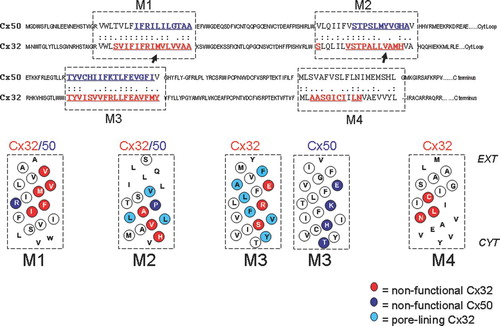
Figure 2 Junctional currents recorded from paired Xenopus oocytes after expression of Cx32 mutants demonstrate that side-chain properties are critical for channel function. Upper traces represent the “reserve-gating” phenotype observed when mutant channels are paired heterotypically with wtCx32. In both cases currents activate when the mutant-expressing oocyte experiences relatively positive transjunctional voltage. Lower traces represent amino substitutions at the same two sites, M34 and V91, which did not significantly alter channel function. At position M34, substitution of amino acids with side chains shorter than the native methionine induced the “reverse-gating” phenotype, whereas side chains that were comparable in length to those of methionine did not disrupt function. At position V91, only substitution of amino acid with β-branched side-chains produced functional channels. Currents were recorded from an oocyte clamped at −40 mV, whereas its partner was pulsed in 10 mV increments (left hand traces) or 20-mV increments (right hand traces) to +60 and −140 mV.
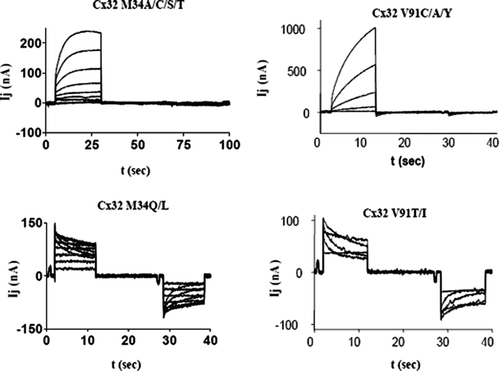
Figure 3 Tentative arrangement of transmembrane helices based on the location of mutation-sensitive sites within the helices. Helical wheel plots are used to show the relative location of substitution-sensitive sites in Cx32 (highlighted with red circles), Cx50 (highlighted with blue circles), as well as substitution-sensitive sites common to both (highlighted with red/blue shading). The two sites in M1 highlighted in red/yellow ovals were shown to be accessible to aqueous thiol reagents added from the outside of intact oocytes, and are thus likely to face a aqueous crevice between helices M1 and M4. For simplicity amino acid numbers correspond to the amino acid positions in Cx32 (see for corresponding Cx50 residues). Helices are arranged as proposed earlier by Skerrett et al. (Citation2002), including the close apposition documented for E146 and C201 in the closed state. This arrangement of helices is in general agreement with the recently published crystal structure of Cx26 (Oshima et al. Citation2007). Protein function was unaffected by tryptophan insertion at positions highlighted in green, suggesting that these residues face a lipid or aqueous environment.
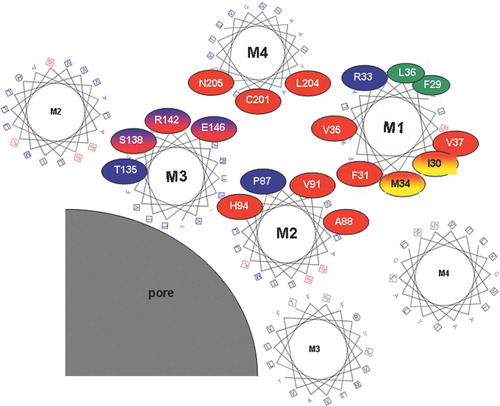
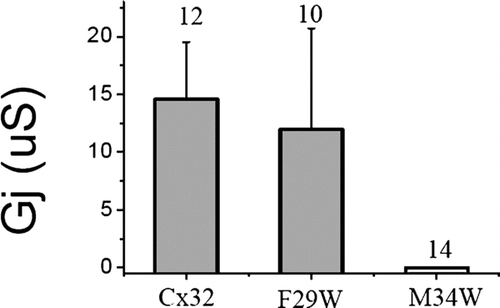
Figure 4 Junctional conductance induced by tryptophan substitution in Cx32 shows no effect at position F29, but a complete loss of function at M34 on the other side of the M1 helix. Each mutant was tested heterotypically with wtCx32 and conductance was determined from the maximum current induced during a +/−100-mV voltage step. Numbers displayed above the bars represent the number of pairs tested. RNA was quantitated by gel electrophoresis and equal amounts of RNA were injected into stage VI oocytes for each mutant, as well as for wtCx32. Oocytes were preinjected with 3 ng of morpholino antisense directed against endogenous Xenopus Cx38 1 or 2 days prior to recording.