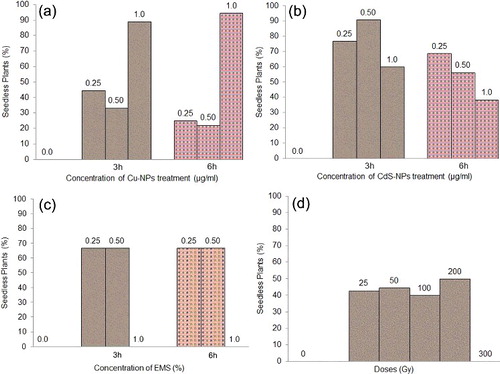
Figures & data
Figure 1. ( a–g) Characterisation of copper and cadmium sulphide nanoparticles. (a–b) UV--vis spectra of (a) Cu-NPs and (b) CdS-NPs. (c) FTIR spectra of CdS-NPs. (d–e) XRD patterns of powdered NPs ((d) Cu; (e) CdS). (f–g) DLS pattern of size distribution of (e) Cu-NPs and (f) CdS- NPs.
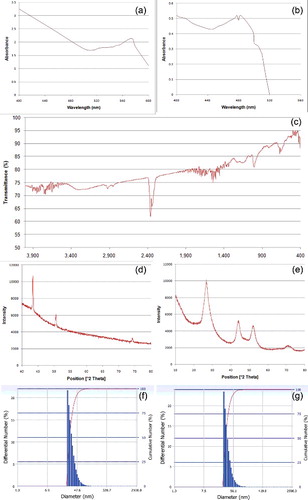
Figure 2. FESEM ((a) Cu-NPs; (b) CdS-NPs) images showing nanoclusters and TEM ((c) Cu-NPs; (d) CdS-NPs) images of NPs (right-handed arrow – gelatin envelop; dotted arrow – metallic core).
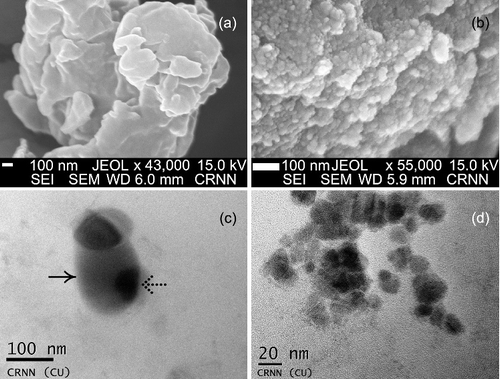
Table 1. Accumulation Cu- and CdS-NPs in controls and treatments.
Figure 3. ( a–g) Mitotic index and frequency of abnormal dividing cells in relation to respective controls of N. sativa. (a–b) Cu-NPs. (c–d) CdS-NPs. (e–f) EMS. (g) Gamma irradiations. (A) Frequency of abnormal cells; (B) mitotic index.
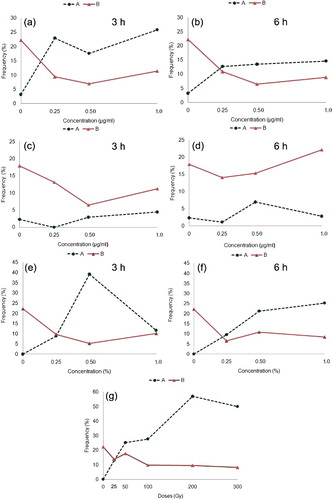
Figure 4. ( a–h) Mitotic cells ((a–e) metaphase, (f–h) anaphase) of N. sativa in (a) control and (b–i) Cu- and CdS-NPs treated material. (a) 2n = 12. (b) Differentially condensed chromosome with a fragment (right-handed arrow) and a ring (dotted arrow). (c) Diplochromatid nature of chromosomes with unoriented chromosomes. (d) Cell with >2n = 12 chromosomes. (e) Differential condensation and fragmentation of chromosomes. (f) Anaphase with lagging fragments. (g) Chromosome bridge formation. (h) I: Anaphasic bridge with two equal size fragment, II: Double bridge with two minute fragments. (i) Cells without or with depleted chromatin. (j) Cell with fragmented chromatin mass. (k) Interphase cell with two unequal sized micronuclei. (l) Giant cell. Scale bar = 20 µm.
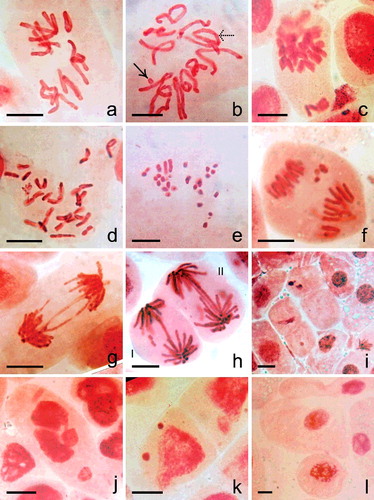
Table 2. Pooled data across doses of treated materials in root tip mitosis of N. sativa.
Table 3. Meiotic configurations and pollen fertility in treated materials.
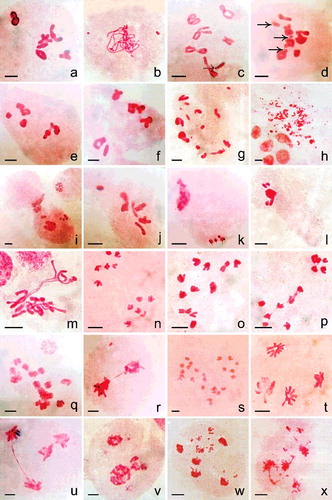
Figure 5. Meiosis in (a) control and (b–x) nanoparticles treated plants. (a) 6II. (b) Pachytene stage with two fragments. (c) Diakinesis cell showing breakage of an arm (dotted arrow) in a bivalent. (d) MI with 5II + 2I + 3 fragments (right-handed arrow). (e–f) Chromosomal grouping (4II + 2II) at MI. (g) Fragments in association to bivalent and univalent at MI possibly 2n > 12. (h) Intense chromosomal fragmentation at MI. (i) Cytomictic behaviour involving two meiocytes. (j) Hyperploid PMC – 7II at MI. (k) Hypoploid cell with condensed type chromosomal fragments. (l) Agglutination of chromatin and fragments. (m) Differential condensation of chromosomes at MI. (n) 6/6 separation at AI. (o) 3/5/4 separation of chromosomes at AI. (p) 5/7 AI separation with interchromosomal connections. (q) 2n > 12 with sticky chromosomes at AI. (r) Dicentric chromatid bridge with an acentric fragment. (s) 2n = 24; AI showing tripolar organisation of chromosomes. (t) AII with one disorganised pole. (u) AII with bridge. (v) Unequal tripolarity. (w,x) AII with multiple groups and fragments. Scale bar = 10 µm.