Figures & data
Figure 1. Overview of the vectors. Above: Map of the starting vectors pB2GW and pB2GV (to scale). The pB2GW vector contains a mini-white gene, a phiC31 integrase compatible attB sequence, a loxP site and an ampicillin resistance (ampR) gene. In pB2GV vector, the screening marker mini-white gene is replaced by vermilion gene. The multiple cloning site (MCS) is flanked by gypsy insulator sequences (In). The starting vectors are suitable for cloning genomic fragments. Below: Schematic of the parental vectors and fluorescent protein (FP) tagging vectors (not to scale). pKW/V and pKW/V-FP are suitable for expression of a gene of interest under the regulation of native promoter and enhancer elements. pUTSW/V, pUTSW/V-FP, pUCSW/V, pUCSW/V-FP, pUPKW/V and pUPKW/V-FP are suitable for Gal4 regulated transgene production. pQUSW/V and pQUSW/V-FP are suitable for QF regulated transgene production. pLASW/V and pLASW/V-FP are suitable for LexA regulated transgene production. pUbSW/V, pUbSW/V-FP, pUbKW/V, pUbKW/V-FP, pT84SW and pT84SW-FP are suitable for ubiquitous expression in germline tissues, embryos, larvae, pupae and adult flies. pT67SW/V and pT67SW/V-FP are suitable for female germline and early embryo expressions. Note that for vermilion version vectors, the XhoI site in MCS cannot be used in vector linearizing because the vermilion sequence contain an XhoI site. K10 pA: K10 polyadenylation signal; SV40 pA: SV40 polyadenylation signal; UAS: Upstream Activation Sequence; hsp70: hsp70 basal promoter; Delta2-3: Delta2-3 transposase promoter; QUAS: QF binding site; LexAop: LexA-binding site; Ubi: Ubiquitin-63E promoter; αTub67C: α-tubulin 67C promoter; αTub84B: α-tubulin 84B promoter; FP: Fluorescent Protein.
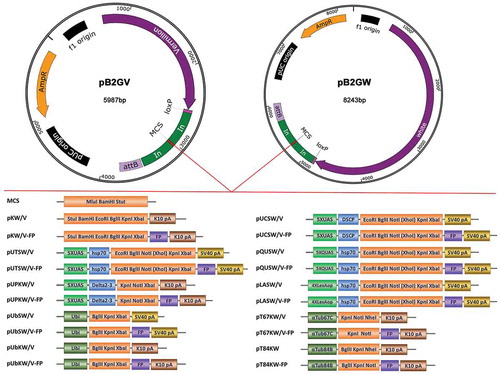
Table 1. Properties and references of the fluorescent proteins.
Table 2. Lethality rescue of dlg5 mutants by dlg5 transgenes.
Figure 2. Expression and localization of the transgene examples. a, Gene structure and mutant alleles of dlg5. Orange boxes represent the coding sequences, and grey boxes represent untranslated regions. The 1.7kb and 9.6kb genomic sequences for Dlg5-EGFP and Dlg5-TagRFP-T constructs are indicated respectively. b and c, Expression and localization of the full-length genomic construct Dlg5-TagRFP-T (b) and the mini genomic construct Dlg5-EGFP (c) in ovaries. d-g, Expression and localization of Ubi-Dlg5.EGFP in egg chamber (D), embryo (e), eye disc (f), and wing disc (g). h-k, Expression and localization of Ubi-Dlg5.TagRFP-T (h), Ubi-Dlg5.mCherry (i), Ubi-CAP.EGFP (j) and Ubi-Corn.EGFP (k) in different stage egg chambers. l-o, Expression and localization of UASt-Dlg5.mCerulean (l), UASt-Dlg5.mCherry (m), UASt-Rack1.EGFP (n, green) and UASt-Corn.Ruby (o) in stage 9 or stage 10 egg chambers. p-q’, Photoactivation of UASt-Dlg5.PAGFP.mRuby and UASt-Dlg5.mEosFP in border cells (p, p’) and follicle cells (q, q’) respectively. The circles (p, p’) or the boxes (q, q’) highlight the region before (Left) and after (Right) the UV laser irradiation in the same sample. ‘pre-active’ (p, q) and ‘post-active’ (p’, q’) are indicated. R and S, Expression and localization of UASt-Lifeact.mOrange and UASt-Lifeact.mClover in S2 Cells. All the UASt transgenes were driven by act5C-Gal4 except the UASt-Rack1.EGFP was driven by slbo-Gal4. Scale bars: 10 um in B-D, F, H-S; 50 um in E and G.
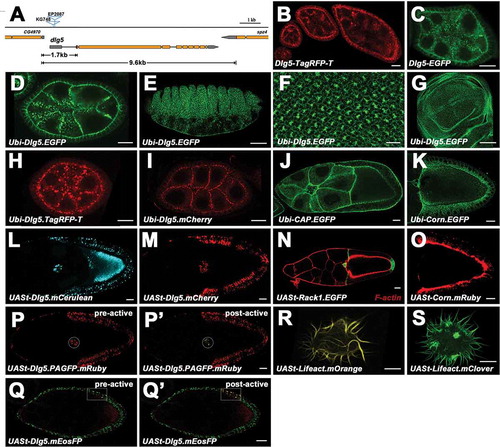
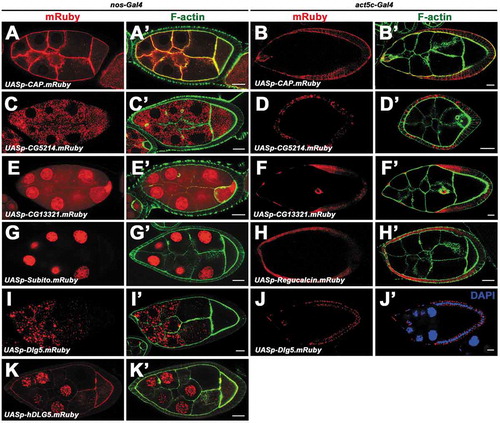
Figure 3. Expression and localization of the UASp transgenes in germline cells and follicle cells. a-k’, Expression and localization of UASp-CAP.mRuby (a-b’), UASp-CG5214.mRuby (c-d’), UASp-CG13321.mRuby (e-f’), UASp-Subito.mRuby (g, g’), UASp-Regucalcin.mRuby (h, h’), UASp-Dlg5.mRuby (i-j’), UASp-hDLG5.mRuby (k, k’) in germline cells (a, a’, c, c’, e, e’, g, g’, i, i’, k, k’) and follicle cells (b, b’, d, d’, f, f’, h, h’, j, j’). nos-Gal4 and act5C-Gal4 was used for germline expression and follicle cell expression respectively. Scale bars: 10um.