Figures & data
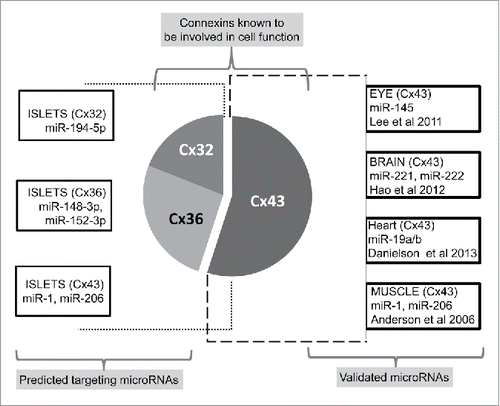
Figure 1. Organization of gap junction proteins. This is a schematic presentation of different types of connexons. A homomeric connexon refers to a connexon formed when 6 identical connexin proteins form the pore for a gap junction. A heteromeric connexon refers to a connexon formed between different connexin subunits. Two identical connexons form a homotypic gap junction, whereas a heterotypic gap junction consists of 2 different hemichannels.
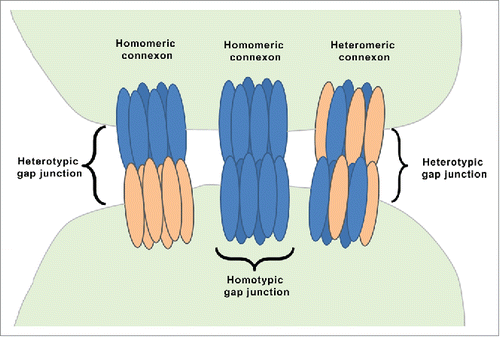
Figure 2. Intercellular transcript regulation via miRNA transfer. miRNAs can enter into neighboring cells via gap junction-mediated transfer. This cartoon demonstrates the ability of a cell (on the left) to transcribe and process the pre-miRNA to mature (single stranded) microRNA. A mature microRNA gets incorporated into RISC (RNA-induced silencing complex) and can target the expression of mRNAs in the same cell via translation inhibition/ transcript degradation or may enter the adjoining cell(s) to inhibit the expression of a specific set of genes in the neighboring cells.
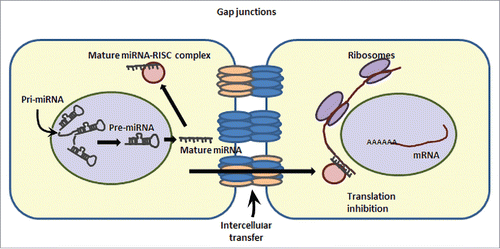
Figure 3. Regulatory microRNAs targeting connexin gene transcripts. Schematic illustrating different microRNAs targeting the expression of specific connexin molecules demonstrated herein. The most widely studied Cx43 is experimentally shown to be targeted by several different miRNAs in various tissues (Right side panel). Left side panel represents different connexins in islets and bioinformatically identified, highly conserved (using the publically available site http://www.targetscan.org) microRNAs predicted to bind to a specific connexin.