Figures & data
Table 1. Description of Proteins
Figure 1. Surface Biolayer Interferometry-based binding of mAbs and bifunctional molecules to spike glycoprotein and to pIgR. Antibodies and antigens are indicated in the graphs. Graphs represent magnitude of response (nm) over time. Association and dissociation are displayed along with fitted curves
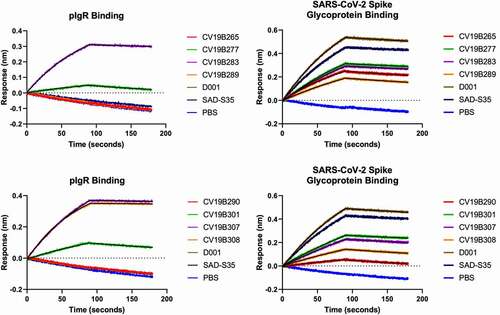
Table 2. Binding kinetic values from surface biolayer interferometry
Table 3. IC50 Values for SARS-CoV-2 competition
Figure 2. Bifunctional molecules Display Specific Functional Activity Against SARS-CoV-2. (a) Functional competition ability is plotted vs. antibody concentration. Antibodies are indicated in the graph. (b) PBMC-mediated ADCC of MDCK-pIgR cells is plotted as green area per well (normalized to 0 h) vs concentration of bifunctional molecule (nM). Antibodies are indicated in the legend
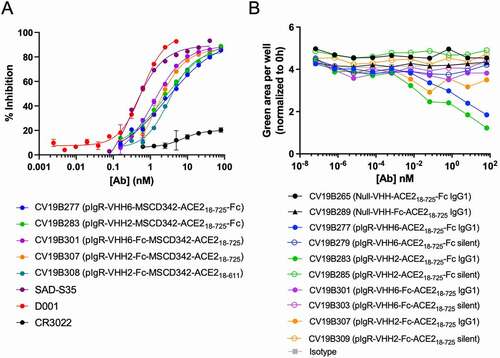
Figure 3. Transcytosis in lung microtissues. A) Transcytosis of bifunctional molecules in EpiAirway tissue model in 24 h post application. For each sample, 20 mg of protein was added to the basolateral well, and after 24 hr, the mucosal surface was washed and the levels of transcytosed antibodies were quantified. Levels are shown in total micrograms transcytosed in a 24 h period. Error bars represent standard error and are representative of at least 2 independent experiments. B-C) Confocal image showing the human EpiAirway microtissue for CV19B307 (b) and CV19B290 (c). Staining shows: blue (nuclei), green (anti-VHH), and red (pIgR). Scale bars shows 50 mm (insets) and 100 mm (main image)
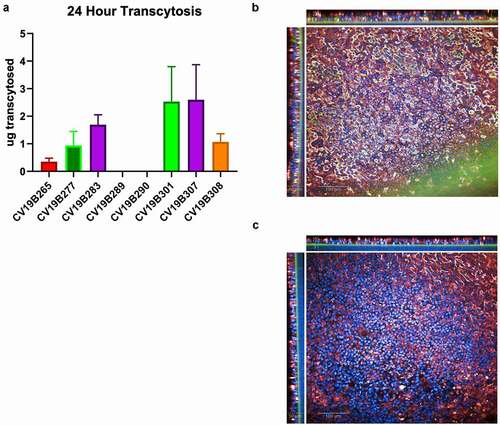
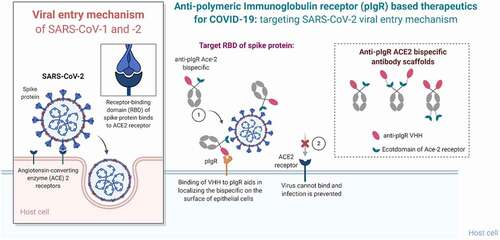
Figure 4. Summary of the mechanism of pIgR-based targeted transport. SARS-CoV-2 viral entry occurs upon binding of the RBD domain of its spike glycoprotein to the ACE2 receptor on target cells. Bifunctional molecules gain access to the lung mucosa through pIgR-mediated transport and bind/neutralize SARS-CoV-2 by binding the RBD domain through their ACE2 ECD moiety in a steric mechanism