Figures & data
Figure 1. LINC01320 is overexpressed in gastric cancer tissues and cells. (a) The expression of LINC01320 in tumor tissues. (b) The expression of LINC01320 expression in normal gastric mucosal cell line and gastric cancer cell lines. (c) Kaplan-Meier curve showed the overall survival in gastric cancer patients according to LINC01320 expression. Red curve represents patients with high LINC01320 expression, while blue curve represents low LINC01320 expression according to the median value of LINC01320. **P< 0.01 vs healthy control or QSG7701
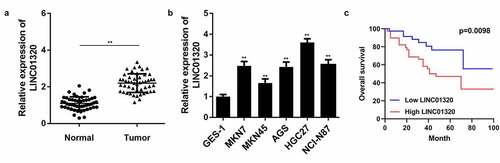
Figure 2. Overexpression of LINC01320 promotes the malignant progression of gastric cancer. (a) Detection of LINC01320 transfection efficiency. (b) After transfected with LINC01320 overexpression or knockdown plasmids, the proliferation ability of gastric cancer cells AGS and HGC27 was detected. (c) After transfected with LINC01320 overexpression or knockdown plasmids,, the migration ability of gastric cancer cells AGS and HGC27 was detected. (d) After transfected with LINC01320 overexpression or knockdown plasmids,, the invasive ability of gastric cancer cells AGS and HGC27 was detected. **P< 0.01 vs sh-nc or vector
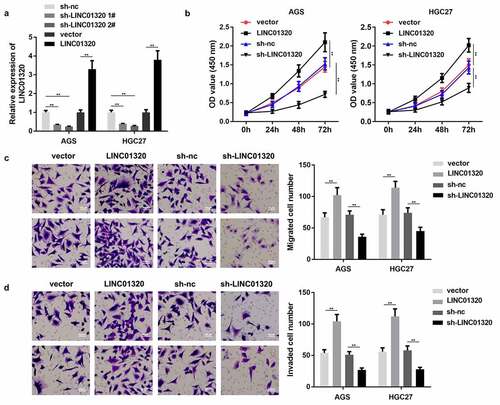
Figure 3. LINC01320 sponges miR-495-5p in cancer cells. (a) The binding sites between LINC01320 and miR-495-5p. (b) Dual luciferase reporter gene detection experiment to verify whether LINC01320 could bind to miR-495-5p. (c) Detection of miR-495-5p expression level. (d) RNA pulled down confirmed the interaction between LINC01320 and miR-495-5p. (e) Detection of miR-495-5p expression in gastric cancer tissues and adjacent tissues. **P< 0.01 vs miR-nc, vector, biotin-nc, normal tissues
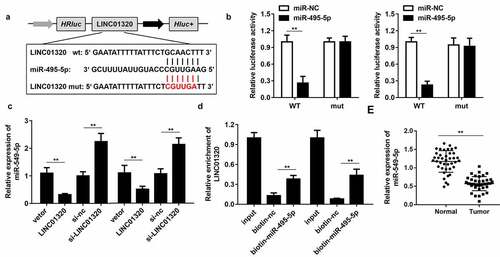
Figure 4. RAB19 is the target gene of miR-495-5p. (a) The binding sites between RAB19 and miR-495-5p. (b) Dual luciferase reporter gene detection experiment to verify whether RAB19 could bind to miR-495-5p. (c) Detection of RAB19 expression level. (d) RNA pulled down confirmed the interaction between RAB19 and miR-495-5p. (e) Detection of RAB19 expression in gastric cancer tissues and adjacent tissues. **P< 0.01 vs miR-nc, vector, biotin-nc, normal tissues
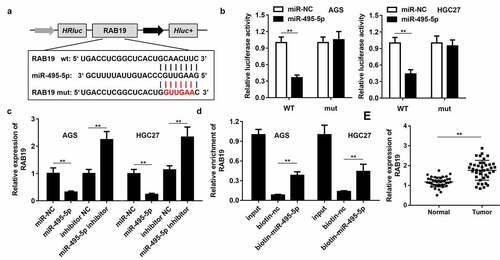
Figure 5. MiR-495-5p overexpression or knockdown of RAB19 reversed the effect of LINC01320 on the proliferation, migration, and invasion of gastric cancer cells. (a) Expression of RAB19 was detected. (b) Cell proliferation experiments after treatment in different groups. (c) Cell migration ability test after different treatments. (d) Cell invasion ability tests after different treatments. **P< 0.01 vs sh-nc, ##P < 0.01 vs sh-LINC01320 + inhibitor nc or sh-LINC01320 + vector
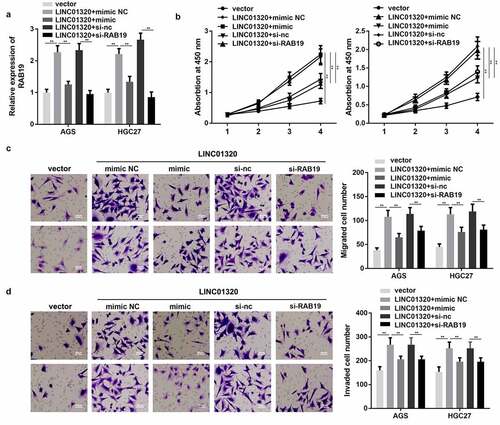
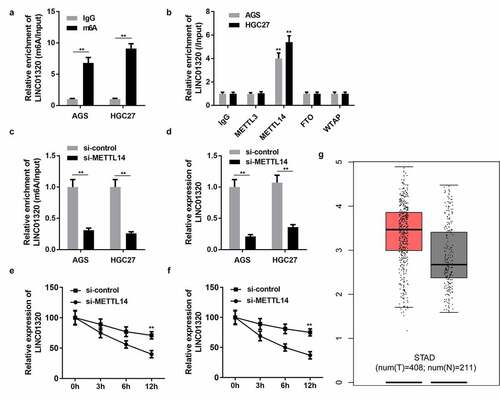
Figure 6. LINC01320 is regulated by m6A modification of METTL14. (a) Protein immunoprecipitation assay to detect the m6A modification of LINC01320. (b) RIP antibody test of different M6A-related proteins. (c) METTL14 interference efficiency detection. (d) LINC01320 expression was detected by qPCR after METTL14 interference. (e and f) METTL14 regulates the m6A modification of LINC01320. **P< 0.01 vs IgG, si-control, or healthy control
Data Availability Statement
All the data is available from the corresponding author due to reasonable request.