Figures & data
Table 1. List of primers used for the plasmid construction and genome integration screening of the transformants. Italic – sequence of overhang for Gibson Assembly®; pBS – pBlueskript II SK (+)
Figure 1. Genetic modification of a PHB overaccumulating mutant Mt_a24. a) Schematic demonstration of genome section of the wild type with the gene exoD and the generated transformant with exchange of exoD through a chloramphenicol resistance gene and the binding sites of the primers for the verification of genomic integration. b) Multiplex-polymerase chain reaction with the OneTaq® Quick Load® DNA polymerase (NEB) and the primers 6–34 F, 10–15 F and 10–16 R to verify the knockout (KO) of exoD in the transformants generated via electroporation of the Mt_a24 mutant
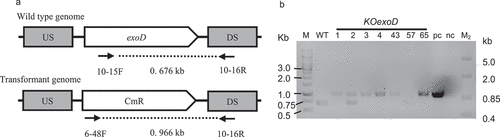
Figure 2. Characterization of the unmodified mutant strain Mt_a24 and its two transformants KOexoD#4 and KOexoD#65 carrying a knockout of the gene exoD by replacement with a chloramphenicol resistance gene, during a cultivation of 7 days in standard BG11 medium followed by a 7 days cultivation within medium deficient of nitrogen and phosphorus with an intermediate sampling on day 4 of limitation. a) determination of the dry cell weight (DCW) by measurement of the optical density at 750 nm; b) analysis of the polyhydroxybutyrate (PHB) yield after acid hydrolyzation and HPLC measurement; c) analysis of the glycogen yield after acid hydrolyzation and IC measurement
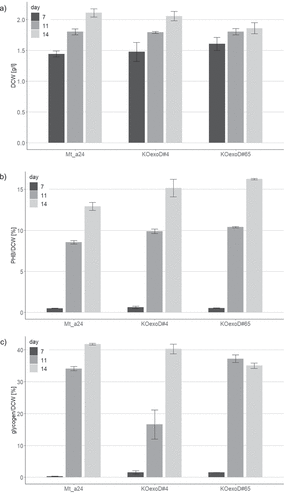
Figure 3. Analysis of the three different EPS fractions, which are either excreted into the medium (S-EPS), loosely bound (LB-EPS) or tightly bound (TB-EPS) to the cell, of the unmodified control strain Mt_a24 and its two transformants KOexoD#4 and KOexoD#65 in standard and nitrogen and phosphorus limited medium (-NP). The analysis was done by hydrolyzation of EPS into monomers, reacted with phenol and measured via a spectrophotometer
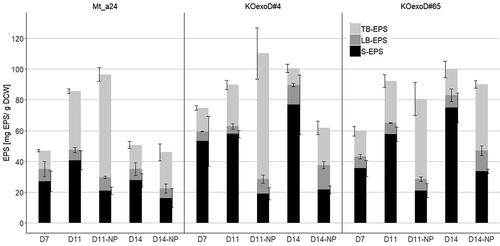
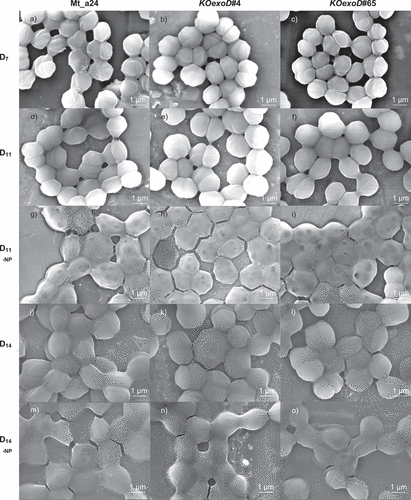
Figure 4. Impact of exoD gene knockout in the two transformants KOexoD#4 (b, e, h, k, n) and KOexoD#65 (c, f, i, l, o) compared to the control strain Mt_a24 (a, d, g, j, m) under different cultivation conditions showed by scanning electron microscope (SEM) imaging. The samples were prepared by fixing the cells (OD730 = 0.7) in glutaraldehyde overnight followed by three washing steps, Au-spattering and imaging at the TESCAN MIRA3 FEG-SEM microscope. Images a – c, standard cultivation conditions on day 7 (D7); images d – f standard cultivation conditions on day 11 (D11); images g – i nitrogen and phosphorus limited medium on day 11 (D11-NP); images j – l standard cultivation conditions on day 14 (D14); images m – o nitrogen and phosphorus limited medium on day 14 (D14-NP). Black marks (D11) on the cells resulted from the measurement. The pattern on the cell surface results from the Au spatter coating