Figures & data
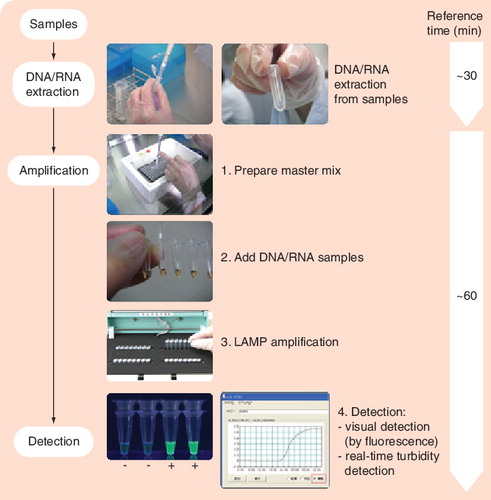
LAMP is characterized by its use of four different primers specifically designed to recognize six distinct regions on the target gene, and its process being performed at a constant temperature using a strand displacement reaction. Image courtesy of Eiken Genome Site, Eiken Chemical Co., Ltd (Tokyo, Japan) and reproduced with permission.
LAMP: Loop-mediated isothermal amplification.
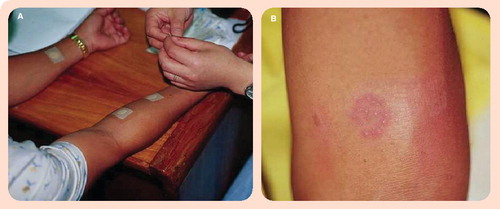
(A) An aqueous solution containing MPB64 protein is applied to the gauze of the Torii patch, and the patch is then immediately applied to the forearm. After 48–72 h, the patch is removed and the response assessed. (B) A positive patch test result. Image courtesy of Sequella, Inc. (MD, USA) and reproduced with permission.
Mycobacterial growth changes the color of the medium from red to yellow. Common contaminants, such as fungi or Gram-negative bacteria, change the color from red to green. Image courtesy of Salubris, Inc. (MA, USA) and reproduced with permission.

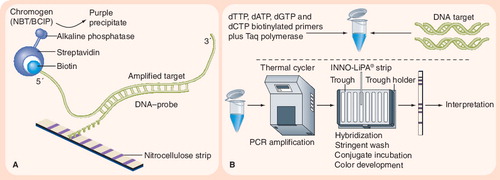
(A) Principle of reverse hybridization. (B) INNO-LiPA® assay. The INNO-LiPA test contains ten oligonucleotide probes (one specific for the Mycobacterium tuberculosis complex, five overlapping wild-type S probes, and four R probes for detecting specific mutations) that are immobilized on nitrocellulose strips. LiPA is performed by extracting DNA and amplifying the rifampicin resistance-determining region of the rpoB gene using PCR. The PCR products are then hybridized with the immobilized probes, and results are determined by colorimetric development. Image adapted from Innogenetics NV (Gent, Belgium) © 2006 Innogenetics Group.
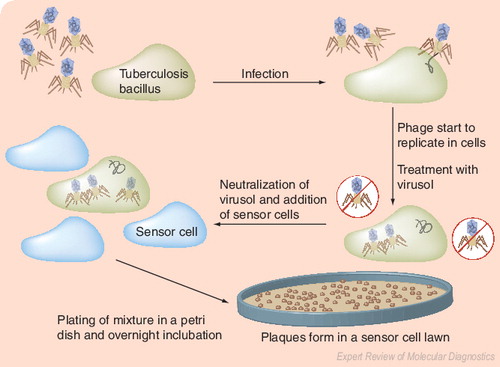
The underlying principle is the amplification of bacteriophages after their infection of M. tuberculosis, followed by detection of progeny phages as lytic plaques on a lawn of Mycobacterium smegmatis. Image courtesy of Biotec Laboratories Ltd (Ipswich, UK), and adapted and reproduced with permission.
The circular disposable is inoculated in the center slot, and is then sealed for the rest of the process. The sample moves into the first six wells, which contain the appropriate antibiotic or control. Using the centrifuge integrated into the main unit, the sample is then moved to the next ring of six wells, which contain luciferase reporter phage. The phage are given time to infect any living mycobacteria, and the sample is then moved into the next ring of six wells, which contain luciferin. Phages that infect living mycobacteria will produce light, which is detected and analyzed by the main Bronx box unit. Image courtesy of Sequella, Inc. (MD, USA) and reproduced with permission.

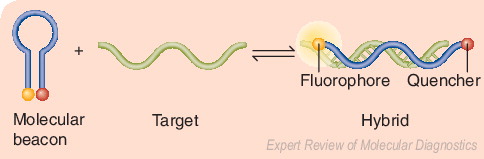
Beacons are single-stranded oligonucleotide hybridization probes that form a stem-and-loop structure. In the absence of the target sequence, the fluorophore and quencher are closely bound, and this prevents fluorescence. When the target sequence is present, hybridization of the probe (loop) with the target results in dissociation of the fluorophore and quencher, thereby restoring fluorescence. Adapted from a figure provided by Sanjay Tyagi.